EcoFOCI evaluation/application of Glider Tools for post processing¶
Summary
Resources
[https://github.com/GliderToolsCommunity/GliderTools](GliderTools Python Package) for secondary processing of data
[https://gliders.ioos.us/data/](IOOS Glider DAC)
[https://underwatergliders.org/index.php/data-management-tools/](UnderWaterGlider Users Group)
Oculus Glider Data Applicability¶
There are pre-existing QC procedures built into both, SeaGlider data (what GliderTools works with) and Oculus data as developed by UW.
There are Engineering parameters and science parameters (including platform instrumentation and additional science instrumentation)
Oculus Glider Data Visualization (PMEL local or FOCI NRT)¶
Kongsberg pilot webpage (immediate information but only for pilots and PI’s)
internal plotly dashboard (NRT / NRT QC) - of binned profile data or timeseries data… quick to plot and look at
internal plotly dashboard (Delayed Mode QC), see gliderTools + IOOS QARTOD + PMEL QC
GliderScope??? https://imos.org.au/gliderscope (gui third party-matlab based runtime-needs admin to install on Mac but also is available for Win)
Other Services and gliders¶
https://gliders.ioos.us/map/
Oculus Glider Distribition¶
Via Internal ERDDAP:
NRT (Pre-existing QC from timeseries files or profile files made by the basestation)
profile files are binned by depth intervals (5m) and may be better hosted as a “gridded” dataset via erddap than tabular
timeseries is one long datastream
Processed and Cleaned
initial qc with improved dials?
GliderTools
IOOS QARTOD QC
Modified Science Routines
BeringSea interface tools
Via IOOS GliderDAC
Examples from BS long mission (2017)¶
Local erddap has two datastreams available for simplicity… the timeseries merged profiles data and the upanddown merged profiles data (from the base station).
Start with gridded (5m) basestation data (NRT level quicklooks with some on base station QC)¶
from erddapy import ERDDAP
import pandas as pd
server_url = 'http://akutan.pmel.noaa.gov:8080/erddap'
e = ERDDAP(server=server_url)
# timeseries or upanddown (profiles)
df = pd.read_csv(e.get_search_url(response='csv', search_for='datasets_sg401_2017_'))
df
| griddap | Subset | tabledap | Make A Graph | wms | files | Title | Summary | FGDC | ISO 19115 | Info | Background Info | RSS | Institution | Dataset ID | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | NaN | http://akutan.pmel.noaa.gov:8080/erddap/tabled... | http://akutan.pmel.noaa.gov:8080/erddap/tabled... | http://akutan.pmel.noaa.gov:8080/erddap/tabled... | NaN | http://akutan.pmel.noaa.gov:8080/erddap/files/... | Physical data collected from Seaglider SG401 d... | PMEL SG401 Bering Sea Test Apr 26, 2017. Up an... | http://akutan.pmel.noaa.gov:8080/erddap/metada... | http://akutan.pmel.noaa.gov:8080/erddap/metada... | http://akutan.pmel.noaa.gov:8080/erddap/info/d... | https://data.nodc.noaa.gov/accession/0092291 | http://akutan.pmel.noaa.gov:8080/erddap/rss/da... | http://akutan.pmel.noaa.gov:8080/erddap/subscr... | NOAA/PMEL | datasets_sg401_2017_upanddown |
| 1 | NaN | http://akutan.pmel.noaa.gov:8080/erddap/tabled... | http://akutan.pmel.noaa.gov:8080/erddap/tabled... | http://akutan.pmel.noaa.gov:8080/erddap/tabled... | NaN | http://akutan.pmel.noaa.gov:8080/erddap/files/... | Physical data collected from Seaglider SG401 d... | PMEL SG401 Bering Sea Test Apr 26, 2017. Times... | http://akutan.pmel.noaa.gov:8080/erddap/metada... | http://akutan.pmel.noaa.gov:8080/erddap/metada... | http://akutan.pmel.noaa.gov:8080/erddap/info/d... | https://data.nodc.noaa.gov/accession/0092291 | http://akutan.pmel.noaa.gov:8080/erddap/rss/da... | http://akutan.pmel.noaa.gov:8080/erddap/subscr... | NOAA/PMEL - EcoFOCI | datasets_sg401_2017_timeseries |
| 2 | http://akutan.pmel.noaa.gov:8080/erddap/gridda... | NaN | NaN | http://akutan.pmel.noaa.gov:8080/erddap/gridda... | NaN | http://akutan.pmel.noaa.gov:8080/erddap/files/... | Physical, chemical, and biological data collec... | SG401 Bering Sea. Applied Physics Lab\n ... | NaN | NaN | http://akutan.pmel.noaa.gov:8080/erddap/info/d... | https://data.nodc.noaa.gov/accession/0092291 | http://akutan.pmel.noaa.gov:8080/erddap/rss/da... | http://akutan.pmel.noaa.gov:8080/erddap/subscr... | Applied Physics Lab University of Washington S... | datasets_sg401_2017_upanddown_gridded |
#thin convenience wrapper for loading FOCI erddap glider data
import GliderDataLoader
sg401=GliderDataLoader.erddap_glider('datasets_sg401_2017_upanddown')
sg401.list_data(verbose=True)
0 datasets_sg401_2017_upanddown
1 datasets_sg401_2017_upanddown_gridded
Name: Dataset ID, dtype: object
variables = {'datasets_sg401_2017_upanddown':['profile_id',
'latitude',
'depth',
'longitude',
'salinity',
'density',
'time',
'temperature']}
sg401.constrain_data(variables=variables,start_date='2017-01-01T00:00:00Z')
dfs = sg401.load_data(year='2017')
datasets_sg401_2017_upanddown
datasets_sg401_2017_upanddown_gridded
Failed to generate url datasets_sg401_2017_upanddown_gridded
sg401.plot_timeseries(dfs['datasets_sg401_2017_upanddown'].sort_index(), 'temperature (degree_C)','depth (m)','temperature (DegC)')
(<Figure size 1224x144 with 2 Axes>, <AxesSubplot:ylabel='Depth (m)'>)
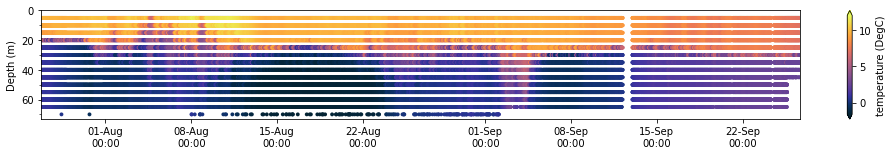
Use Gridded ERRDAP Dataset if Available for binned data¶
#all
import cmocean
e.protocol = 'griddap'
e.dataset_id='datasets_sg401_2017_upanddown_gridded'
e.variables = ['temperature','ctd_time']
e.response = 'ncCF'
ds = e.to_xarray(decode_times=True)
ds.temperature.T.plot(yincrease=False,cmap=cmocean.cm.thermal,vmin=-2,vmax=12,figsize=(18,2))
<matplotlib.collections.QuadMesh at 0x7fb4dbbc5fa0>
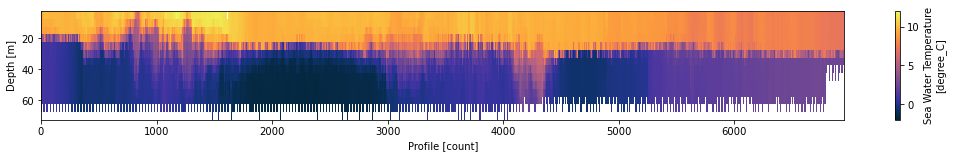
Now using larger archived timeseries data ((NRT level quicklooks with some on base station QC)¶
sg401=GliderDataLoader.erddap_glider('datasets_sg401_2017_timeseries')
sg401.list_data(verbose=True)
0 datasets_sg401_2017_timeseries
Name: Dataset ID, dtype: object
variables = {'datasets_sg401_2017_timeseries':['profile_id',
'latitude',
'depth',
'longitude',
'salinity',
'density',
'time',
'temperature']}
sg401.constrain_data(variables=variables,start_date='2017-01-01T00:00:00Z')
dfs = sg401.load_data(year='2017')
datasets_sg401_2017_timeseries
sg401.plot_timeseries(dfs['datasets_sg401_2017_timeseries'].sort_index(), 'temperature (degree_C)','depth (m)','temperature (DegC)')
(<Figure size 1224x144 with 2 Axes>, <AxesSubplot:ylabel='Depth (m)'>)
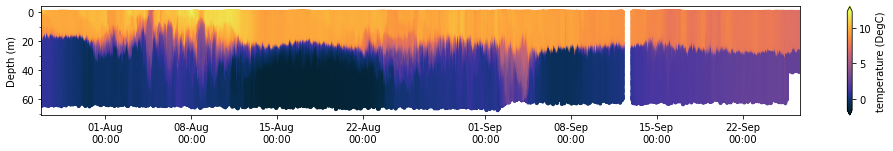
Loading individual dive datasets and running GliderTools¶
This isn’t truly Delayed mode data (just delayed mode processing)
%load_ext autoreload
# pylab for more MATLAB like environment and inline displays plots below cells
%pylab inline
# if gsw Warning shows, manually install gsw if possible - will still work without
import glidertools as gt
from cmocean import cm as cmo # we use this for colormaps
Populating the interactive namespace from numpy and matplotlib
Loading data¶
Working with Seaglider base station files¶
GliderTools supports loading Seaglider files, including scicon data (different sampling frequencies).
There is a function that makes it easier to find variable names that you’d like to load: gt.load.seaglider_show_variables
This function is demonstrated in the cell below.
The function accepts a list of file names and can also receive a string with a wildcard placeholder (*) and basic regular expressions are also supported. In the example below we use a simple asterisk placeholder for all the files.
Note that the function chooses only one file from the passed list or glob string - this file name will be shown. The returned table shows the variable name, dimensions, units and brief comment if it is available.
#skipp first 1000 dives
filenames = 'test_data/sg401_2017/p401[123]*.nc'
gt.load.seaglider_show_variables(filenames)
information is based on file: test_data/sg401_2017/p4012283.nc
| dims | units | comment | |
|---|---|---|---|
| name | |||
| gc_data_pts | gc_event | 1 | |
| gc_depth | gc_event | meters | |
| gc_end_secs | gc_event | seconds since 1970-1-1 00:00:00 | End of GC time in GMT epoch format |
| gc_gcphase | gc_event | ||
| gc_humidity | gc_event | percent | Internal relative humidity |
| gc_int_press | gc_event | psia | Internal pressure |
| gc_ob_vertv | gc_event | cm/s | |
| gc_pitch_ad | gc_event | 1 | |
| gc_pitch_ad_start | gc_event | 1 | |
| gc_pitch_ctl | gc_event | cm | |
| gc_pitch_errors | gc_event | 1 | |
| gc_pitch_i | gc_event | A | |
| gc_pitch_retries | gc_event | 1 | |
| gc_pitch_secs | gc_event | seconds | Elapsed seconds since start of this pitch change |
| gc_pitch_volts | gc_event | V | |
| gc_roll_ad | gc_event | 1 | |
| gc_roll_ad_start | gc_event | 1 | |
| gc_roll_errors | gc_event | 1 | |
| gc_roll_i | gc_event | A | |
| gc_roll_retries | gc_event | 1 | |
| gc_roll_secs | gc_event | seconds | Elapsed seconds since start of this roll change |
| gc_roll_volts | gc_event | V | |
| gc_st_secs | gc_event | seconds since 1970-1-1 00:00:00 | Start of GC time in GMT epoch format |
| gc_vbd_ad | gc_event | 1 | |
| gc_vbd_ctl | gc_event | cm | |
| gc_vbd_errors | gc_event | 1 | |
| gc_vbd_i | gc_event | A | |
| gc_vbd_pot1_ad | gc_event | 1 | |
| gc_vbd_pot1_ad_start | gc_event | 1 | |
| gc_vbd_pot2_ad | gc_event | 1 | |
| gc_vbd_pot2_ad_start | gc_event | 1 | |
| gc_vbd_retries | gc_event | 1 | |
| gc_vbd_secs | gc_event | seconds | Elapsed seconds since start of this VBD change |
| gc_vbd_volts | gc_event | V | |
| log_gps_lat | gps_info | degrees_north | GPS latitudes |
| log_gps_lon | gps_info | degrees_east | GPS longitudes |
| log_gps_time | gps_info | seconds since 1970-1-1 00:00:00 | GPS times in GMT epoch format |
| eng_mag_x | magnetometer_data_point | magnetometer x | |
| eng_mag_y | magnetometer_data_point | magnetometer y | |
| eng_mag_z | magnetometer_data_point | magnetometer z | |
| eng_satd_PARuV | sat1000_data_point | ||
| eng_satd_Temp | sat1000_data_point | ||
| eng_satd_Timer | sat1000_data_point | ||
| eng_satd_pitch | sat1000_data_point | ||
| eng_satd_roll | sat1000_data_point | ||
| eng_satu_PARuV | sat1001_data_point | ||
| eng_satu_Temp | sat1001_data_point | ||
| eng_satu_Timer | sat1001_data_point | ||
| eng_satu_pitch | sat1001_data_point | ||
| eng_satu_roll | sat1001_data_point | ||
| aanderaa4330_dissolved_oxygen | sg_data_point | micromoles/kg | Dissolved oxygen concentration, calculated from optode tcphase corrected for salininty and depth |
| aanderaa4330_dissolved_oxygen_qc | sg_data_point | Whether to trust each optode dissolved oxygen value | |
| aanderaa4330_instrument_dissolved_oxygen | sg_data_point | micromoles/kg | Dissolved oxygen concentration reported from optode corrected for salinity |
| aanderaa4330_results_time | sg_data_point | seconds since 1970-1-1 00:00:00 | time for Aanderaa 4330 in GMT epoch format |
| buoyancy | sg_data_point | g | Buoyancy of vehicle, corrected for compression effects |
| conductivity | sg_data_point | S/m | Conductivity corrected for anomalies |
| conductivity_qc | sg_data_point | Whether to trust each corrected conductivity value | |
| conductivity_raw | sg_data_point | S/m | Uncorrected conductivity |
| conductivity_raw_qc | sg_data_point | Whether to trust each raw conductivity value | |
| ctd_depth | sg_data_point | meters | CTD thermistor depth corrected for average latitude |
| ctd_pressure | sg_data_point | dbar | Pressure at CTD thermistor |
| ctd_time | sg_data_point | seconds since 1970-1-1 00:00:00 | Time of CTD sample in GMT epoch format |
| density | sg_data_point | g/m^3 | Sea water potential density |
| density_insitu | sg_data_point | g/m^3 | Sea water in-situ density based on pressure |
| depth | sg_data_point | meters | Depth below the surface, corrected for average latitude |
| dissolved_oxygen_sat | sg_data_point | micromoles/kg | Calculated saturation value for oxygen given measured presure and corrected temperature, and salinity |
| east_displacement | sg_data_point | meters | Eastward displacement from hdm |
| east_displacement_gsm | sg_data_point | meters | Eastward displacement from gsm |
| eng_GC_phase | sg_data_point | ||
| eng_aa4330_AirSat | sg_data_point | As reported by the instrument | |
| eng_aa4330_CalPhase | sg_data_point | As reported by the instrument | |
| eng_aa4330_O2 | sg_data_point | micromoles/L | Dissolved oxygen as reported by the instument, based on on-board calibration data, assuming optode temperature but without depth or salinity correction |
| eng_aa4330_TCPhase | sg_data_point | As reported by the instrument | |
| eng_aa4330_Temp | sg_data_point | degrees_Celsius | As reported by the instrument |
| eng_depth | sg_data_point | cm | Measured vertical distance below the surface |
| eng_elaps_t | sg_data_point | seconds | Elapsed seconds since start of dive |
| eng_elaps_t_0000 | sg_data_point | seconds | Elapsed seconds since start of mission |
| eng_head | sg_data_point | degrees | Vehicle heading (magnetic) |
| eng_pitchAng | sg_data_point | degrees | Vehicle pitch |
| eng_pitchCtl | sg_data_point | ||
| eng_rec | sg_data_point | ||
| eng_rollAng | sg_data_point | degrees | Vehicle roll |
| eng_rollCtl | sg_data_point | ||
| eng_sbect_condFreq | sg_data_point | As reported by the instrument | |
| eng_sbect_tempFreq | sg_data_point | As reported by the instrument | |
| eng_vbdCC | sg_data_point | ||
| eng_wlbb2fl_sig470nm | sg_data_point | blue scattering as reported by instrument | |
| eng_wlbb2fl_sig695nm | sg_data_point | chlorophyll fluorescence as reported by instrument | |
| eng_wlbb2fl_sig700nm | sg_data_point | red scattering as reported by instrument | |
| eng_wlbb2fl_temp | sg_data_point | temperature as reported by instrument | |
| glide_angle | sg_data_point | cm/s | Glide angle based on hdm |
| glide_angle_gsm | sg_data_point | cm/s | Glide angle based on gsm |
| horz_speed | sg_data_point | cm/s | Vehicle horizontal speed based on hdm |
| horz_speed_gsm | sg_data_point | cm/s | Vehicle horizontal speed based on gsm |
| latitude | sg_data_point | degrees_north | Latitude of the sample based on hdm DAC |
| latitude_gsm | sg_data_point | degrees_north | Latitude of the sample based on gsm DAC |
| longitude | sg_data_point | degrees_east | Longitude of the sample based on hdm DAC |
| longitude_gsm | sg_data_point | degrees_east | Longitude of the sample based on gsm DAC |
| north_displacement | sg_data_point | meters | Northward displacement from hdm |
| north_displacement_gsm | sg_data_point | meters | Northward displacement from gsm |
| pressure | sg_data_point | dbar | Uncorrected sea-water pressure at pressure sensor |
| salinity | sg_data_point | 1e-3 | Salinity corrected for thermal-inertia effects (PSU) |
| salinity_qc | sg_data_point | Whether to trust each corrected salinity value | |
| salinity_raw | sg_data_point | 1e-3 | Uncorrected salinity derived from temperature_raw and conductivity_raw (PSU) |
| salinity_raw_qc | sg_data_point | Whether to trust each raw salinity value | |
| sigma_t | sg_data_point | g/m^3 | Sigma based on density |
| sigma_theta | sg_data_point | g/m^3 | |
| sound_velocity | sg_data_point | m/s | Sound velocity |
| speed | sg_data_point | cm/s | Vehicle speed based on hdm |
| speed_gsm | sg_data_point | cm/s | Vehicle speed based on gsm |
| speed_qc | sg_data_point | Whether to trust each hdm speed value | |
| temperature | sg_data_point | degrees_Celsius | Termperature (in situ) corrected for thermistor first-order lag |
| temperature_qc | sg_data_point | Whether to trust each corrected temperature value | |
| temperature_raw | sg_data_point | degrees_Celsius | Uncorrected temperature (in situ) |
| temperature_raw_qc | sg_data_point | Whether to trust each raw temperature value | |
| theta | sg_data_point | degrees_Celsius | Potential temperature based on corrected salinity |
| time | sg_data_point | seconds since 1970-1-1 00:00:00 | Time of the sample in GMT epoch format |
| vert_speed | sg_data_point | cm/s | Vehicle vertical speed based on hdm |
| vert_speed_gsm | sg_data_point | cm/s | Vehicle vertical speed based on gsm |
| wlbb2fl_results_time | sg_data_point | seconds since 1970-1-1 00:00:00 | wlbb2fl result time in GMT epoch format |
| wlbb2fl_sig470nm_adjusted | sg_data_point | meter^-1 steradian^-1 | total volume blue scattering coefficient using manufacturer-supplied dark counts and scaling factor |
| wlbb2fl_sig695nm_adjusted | sg_data_point | micrograms/liter | chlorophyll-a concentration using manufacturer-supplied dark counts and scaling factor based on phytoplankton monoculture |
| wlbb2fl_sig700nm_adjusted | sg_data_point | meter^-1 steradian^-1 | total volume red scattering coefficient using manufacturer-supplied dark counts and scaling factor |
| CTD_qc | string | Whether to trust the corrected CTD values | |
| GPS1_qc | string | Whether to trust the GPS1 information | |
| GPS2_qc | string | Whether to trust the GPS2 information | |
| GPSE_qc | string | Whether to trust the final GPS information | |
| aa4330 | string | ||
| aanderaa4330_qc | string | Whether to trust the Aanderaa 3880 results | |
| avg_latitude | string | degrees_north | The average latitude of the dive |
| depth_avg_curr_east | string | m/s | Eastward component of the dive depth-average current based on hdm |
| depth_avg_curr_east_gsm | string | m/s | Eastward component of depth-average current based on gsm |
| depth_avg_curr_north | string | m/s | Northward component of the dive depth-average current based on hdm |
| depth_avg_curr_north_gsm | string | m/s | Northward component of depth-average current based on gsm |
| depth_avg_curr_qc | string | Whether to trust the dive depth-average current values and displacements | |
| flight_avg_speed_east | string | m/s | Eastward component of flight average speed based on hdm |
| flight_avg_speed_east_gsm | string | m/s | Eastward component of flight average speed based on gsm |
| flight_avg_speed_north | string | m/s | Northward component of flight average speed based on hdm |
| flight_avg_speed_north_gsm | string | m/s | Northward component of flight average speed based on gsm |
| glider | string | ||
| hdm_qc | string | Whether corrected temperatures, salinities, and velocities from the hydrodynamic model converged on a consistent solution | |
| latlong_qc | string | Whether to trust the dive estimated latitude and longitude estimates | |
| log_10V_AH | string | ||
| log_24V_AH | string | ||
| log_AD7714Ch0Gain | string | ||
| log_AH0_10V | string | ||
| log_AH0_24V | string | ||
| log_ALTIM_BOTTOM_PING_RANGE | string | ||
| log_ALTIM_BOTTOM_TURN_MARGIN | string | ||
| log_ALTIM_FREQUENCY | string | ||
| log_ALTIM_PING_DELTA | string | ||
| log_ALTIM_PING_DEPTH | string | ||
| log_ALTIM_PULSE | string | ||
| log_ALTIM_SENSITIVITY | string | ||
| log_ALTIM_TOP_MIN_OBSTACLE | string | ||
| log_ALTIM_TOP_PING_RANGE | string | ||
| log_ALTIM_TOP_TURN_MARGIN | string | ||
| log_APOGEE_PITCH | string | ||
| log_CALL_NDIVES | string | ||
| log_CALL_TRIES | string | ||
| log_CALL_WAIT | string | ||
| log_CAPMAXSIZE | string | ||
| log_CAPUPLOAD | string | ||
| log_CAP_FILE_SIZE | string | ||
| log_CF8_MAXERRORS | string | ||
| log_CFSIZE | string | ||
| log_COMM_SEQ | string | ||
| log_COMPASS2_DEVICE | string | ||
| log_COMPASS_DEVICE | string | ||
| log_COMPASS_USE | string | ||
| log_COURSE_BIAS | string | ||
| log_C_PITCH | string | ||
| log_C_ROLL_CLIMB | string | ||
| log_C_ROLL_DIVE | string | ||
| log_C_VBD | string | ||
| log_DATA_FILE_SIZE | string | ||
| log_DBDW | string | ||
| log_DEEPGLIDER | string | ||
| log_DEVICE1 | string | ||
| log_DEVICE2 | string | ||
| log_DEVICE3 | string | ||
| log_DEVICE4 | string | ||
| log_DEVICE5 | string | ||
| log_DEVICE6 | string | ||
| log_DEVICES | string | ||
| log_DEVICE_MAMPS | string | ||
| log_DEVICE_SECS | string | ||
| log_DIVE | string | ||
| log_D_ABORT | string | ||
| log_D_BOOST | string | ||
| log_D_CALL | string | ||
| log_D_FINISH | string | ||
| log_D_FLARE | string | ||
| log_D_GRID | string | ||
| log_D_NO_BLEED | string | ||
| log_D_OFFGRID | string | ||
| log_D_PITCH | string | ||
| log_D_SAFE | string | ||
| log_D_SURF | string | ||
| log_D_TGT | string | ||
| log_ERRORS | string | ||
| log_ESCAPE_HEADING | string | ||
| log_ESCAPE_HEADING_DELTA | string | ||
| log_FERRY_MAX | string | ||
| log_FG_AHR_10V | string | ||
| log_FG_AHR_10Vo | string | ||
| log_FG_AHR_24V | string | ||
| log_FG_AHR_24Vo | string | ||
| log_FILEMGR | string | ||
| log_FIX_MISSING_TIMEOUT | string | ||
| log_GLIDE_SLOPE | string | ||
| log_GPS | string | String reported in logfile for GPS fix (first surface position after dive) | |
| log_GPS1 | string | String reported in logfile for GPS1 fix (first surface position before dive) | |
| log_GPS2 | string | String reported in logfile for GPS2 fix (last surface position before dive) | |
| log_GPS_DEVICE | string | ||
| log_HD_A | string | Hydrodynamic lift factor for given hull shape (1/degrees of attack angle) | |
| log_HD_B | string | Hydrodynamic drag factor for given hull shape (Pa^(-1/4)) | |
| log_HD_C | string | Hydrodynamic induced drag factor for given hull shape (1/degrees^2 of attack angle) | |
| log_HEADING | string | ||
| log_HEAD_ERRBAND | string | ||
| log_HEAPDBG | string | ||
| log_HUMID | string | ||
| log_ICE_FREEZE_MARGIN | string | ||
| log_ID | string | ||
| log_INTERNAL_PRESSURE | string | ||
| log_INT_PRESSURE_SLOPE | string | ||
| log_INT_PRESSURE_YINT | string | ||
| log_IRIDIUM_FIX | string | ||
| log_KALMAN_CONTROL | string | ||
| log_KALMAN_USE | string | ||
| log_KALMAN_X | string | ||
| log_KALMAN_Y | string | ||
| log_LOGGERDEVICE1 | string | ||
| log_LOGGERDEVICE2 | string | ||
| log_LOGGERDEVICE3 | string | ||
| log_LOGGERDEVICE4 | string | ||
| log_LOGGERS | string | ||
| log_MASS | string | ||
| log_MASS_COMP | string | ||
| log_MAXI_10V | string | ||
| log_MAXI_24V | string | ||
| log_MAX_BUOY | string | ||
| log_MEM | string | ||
| log_MHEAD_RNG_PITCHd_Wd | string | ||
| log_MINV_10V | string | ||
| log_MINV_24V | string | ||
| log_MISSION | string | ||
| log_MOTHERBOARD | string | ||
| log_NAV_MODE | string | ||
| log_NOCOMM_ACTION | string | ||
| log_N_DIVES | string | ||
| log_N_FILEKB | string | ||
| log_N_GPS | string | ||
| log_N_NOCOMM | string | ||
| log_N_NOSURFACE | string | ||
| log_PHONE_DEVICE | string | ||
| log_PHONE_SUPPLY | string | ||
| log_PITCH_ADJ_DBAND | string | ||
| log_PITCH_ADJ_GAIN | string | ||
| log_PITCH_AD_RATE | string | ||
| log_PITCH_CNV | string | ||
| log_PITCH_DBAND | string | ||
| log_PITCH_GAIN | string | ||
| log_PITCH_MAX | string | ||
| log_PITCH_MAXERRORS | string | ||
| log_PITCH_MIN | string | ||
| log_PITCH_TIMEOUT | string | ||
| log_PITCH_VBD_SHIFT | string | ||
| log_PITCH_W_DBAND | string | ||
| log_PITCH_W_GAIN | string | ||
| log_PRESSURE_SLOPE | string | ||
| log_PRESSURE_YINT | string | ||
| log_PROTOCOL | string | ||
| log_P_OVSHOOT | string | ||
| log_P_OVSHOOT_WITHG | string | ||
| log_RAFOS_CORR_THRESH | string | ||
| log_RAFOS_DEVICE | string | ||
| log_RAFOS_HIT_WINDOW | string | ||
| log_RAFOS_MMODEM | string | ||
| log_RAFOS_PEAK_OFFSET | string | ||
| log_RELAUNCH | string | ||
| log_RHO | string | gram/cc | Expected density at deepest point over the deployment |
| log_ROLL_ADJ_DBAND | string | ||
| log_ROLL_ADJ_GAIN | string | ||
| log_ROLL_AD_RATE | string | ||
| log_ROLL_CNV | string | ||
| log_ROLL_DEG | string | ||
| log_ROLL_MAX | string | ||
| log_ROLL_MAXERRORS | string | ||
| log_ROLL_MIN | string | ||
| log_ROLL_TIMEOUT | string | ||
| log_R_PORT_OVSHOOT | string | ||
| log_R_STBD_OVSHOOT | string | ||
| log_SEABIRD_C_G | string | ||
| log_SEABIRD_C_H | string | ||
| log_SEABIRD_C_I | string | ||
| log_SEABIRD_C_J | string | ||
| log_SEABIRD_C_Z | string | Conductivity zero frequency | |
| log_SEABIRD_T_G | string | ||
| log_SEABIRD_T_H | string | ||
| log_SEABIRD_T_I | string | ||
| log_SEABIRD_T_J | string | ||
| log_SENSORS | string | ||
| log_SENSOR_MAMPS | string | ||
| log_SENSOR_SECS | string | ||
| log_SIM_PITCH | string | ||
| log_SIM_W | string | ||
| log_SM_CC | string | ||
| log_SM_CCo | string | ||
| log_SM_GC | string | ||
| log_SPEED_FACTOR | string | ||
| log_SPEED_LIMITS | string | ||
| log_STOP_T | string | ||
| log_STROBE | string | ||
| log_SURFACE_URGENCY | string | ||
| log_SURFACE_URGENCY_FORCE | string | ||
| log_SURFACE_URGENCY_TRY | string | ||
| log_TCM_PITCH_OFFSET | string | ||
| log_TCM_ROLL_OFFSET | string | ||
| log_TCM_TEMP | string | ||
| log_TGT_AUTO_DEFAULT | string | ||
| log_TGT_DEFAULT_LAT | string | ||
| log_TGT_DEFAULT_LON | string | ||
| log_TGT_LATLONG | string | ||
| log_TGT_NAME | string | ||
| log_TGT_RADIUS | string | ||
| log_TT8_MAMPS | string | ||
| log_T_ABORT | string | ||
| log_T_BOOST | string | ||
| log_T_DIVE | string | ||
| log_T_EPIRB | string | ||
| log_T_GPS | string | ||
| log_T_LOITER | string | ||
| log_T_MISSION | string | ||
| log_T_NO_W | string | ||
| log_T_RSLEEP | string | ||
| log_T_SLOITER | string | ||
| log_T_TURN | string | ||
| log_T_TURN_SAMPINT | string | ||
| log_T_WATCHDOG | string | ||
| log_UNCOM_BLEED | string | ||
| log_UPLOAD_DIVES_MAX | string | ||
| log_USE_BATHY | string | ||
| log_USE_ICE | string | ||
| log_VBD_BLEED_AD_RATE | string | ||
| log_VBD_CNV | string | ||
| log_VBD_DBAND | string | ||
| log_VBD_LP_IGNORE | string | ||
| log_VBD_MAX | string | ||
| log_VBD_MAXERRORS | string | ||
| log_VBD_MIN | string | ||
| log_VBD_PUMP_AD_RATE_APOGEE | string | ||
| log_VBD_PUMP_AD_RATE_SURFACE | string | ||
| log_VBD_TIMEOUT | string | ||
| log_W_ADJ_DBAND | string | ||
| log_XPDR_DEVICE | string | ||
| log_XPDR_INHIBIT | string | ||
| log_XPDR_PINGS | string | ||
| log_XPDR_VALID | string | ||
| log__CALLS | string | ||
| log__SM_ANGLEo | string | ||
| log__SM_DEPTHo | string | ||
| log__XMS_NAKs | string | ||
| log__XMS_TOUTs | string | ||
| magnetic_variation | string | The magnetic variance from true north (degrees) | |
| magnetometer | string | ||
| reviewed | string | Whether a scientist has reviewed and approved this profile | |
| sat1000 | string | ||
| sat1001 | string | ||
| sbe41 | string | ||
| sg_cal_abs_compress | string | cc/dbar | SG vehicle compressibility |
| sg_cal_c_g | string | ||
| sg_cal_c_h | string | ||
| sg_cal_c_i | string | ||
| sg_cal_c_j | string | ||
| sg_cal_calibcomm | string | ||
| sg_cal_calibcomm_optode | string | ||
| sg_cal_calibcomm_wetlabs | string | ||
| sg_cal_comm_oxy_type | string | ||
| sg_cal_cpcor | string | Nominal compression factor of conductivity tube with pressure | |
| sg_cal_ctcor | string | Nominal thermal expansion factor of a cube of boro-silicate glass | |
| sg_cal_hd_a | string | Hydrodynamic lift factor for given hull shape (1/degrees of attack angle) | |
| sg_cal_hd_b | string | Hydrodynamic drag factor for given hull shape (Pa^(-1/4)) | |
| sg_cal_hd_c | string | Hydrodynamic induced drag factor for given hull shape (1/radians^2 of attack angle) | |
| sg_cal_id_str | string | Three digit vehicle identification string | |
| sg_cal_mass | string | kg | Mass of the glider |
| sg_cal_mass_comp | string | kg | Mass of the compressee |
| sg_cal_mission_title | string | Description of mission | |
| sg_cal_optode_ConcCoef0 | string | ||
| sg_cal_optode_ConcCoef1 | string | ||
| sg_cal_optode_FoilCoefA0 | string | ||
| sg_cal_optode_FoilCoefA1 | string | ||
| sg_cal_optode_FoilCoefA10 | string | ||
| sg_cal_optode_FoilCoefA11 | string | ||
| sg_cal_optode_FoilCoefA12 | string | ||
| sg_cal_optode_FoilCoefA13 | string | ||
| sg_cal_optode_FoilCoefA2 | string | ||
| sg_cal_optode_FoilCoefA3 | string | ||
| sg_cal_optode_FoilCoefA4 | string | ||
| sg_cal_optode_FoilCoefA5 | string | ||
| sg_cal_optode_FoilCoefA6 | string | ||
| sg_cal_optode_FoilCoefA7 | string | ||
| sg_cal_optode_FoilCoefA8 | string | ||
| sg_cal_optode_FoilCoefA9 | string | ||
| sg_cal_optode_FoilCoefB0 | string | ||
| sg_cal_optode_FoilCoefB1 | string | ||
| sg_cal_optode_FoilCoefB10 | string | ||
| sg_cal_optode_FoilCoefB11 | string | ||
| sg_cal_optode_FoilCoefB12 | string | ||
| sg_cal_optode_FoilCoefB13 | string | ||
| sg_cal_optode_FoilCoefB2 | string | ||
| sg_cal_optode_FoilCoefB3 | string | ||
| sg_cal_optode_FoilCoefB4 | string | ||
| sg_cal_optode_FoilCoefB5 | string | ||
| sg_cal_optode_FoilCoefB6 | string | ||
| sg_cal_optode_FoilCoefB7 | string | ||
| sg_cal_optode_FoilCoefB8 | string | ||
| sg_cal_optode_FoilCoefB9 | string | ||
| sg_cal_optode_PhaseCoef0 | string | ||
| sg_cal_optode_PhaseCoef1 | string | ||
| sg_cal_optode_PhaseCoef2 | string | ||
| sg_cal_optode_PhaseCoef3 | string | ||
| sg_cal_optode_SVUCoef0 | string | ||
| sg_cal_optode_SVUCoef1 | string | ||
| sg_cal_optode_SVUCoef2 | string | ||
| sg_cal_optode_SVUCoef3 | string | ||
| sg_cal_optode_SVUCoef4 | string | ||
| sg_cal_optode_SVUCoef5 | string | ||
| sg_cal_optode_SVUCoef6 | string | ||
| sg_cal_optode_SVU_enabled | string | ||
| sg_cal_rho0 | string | kg/m^3 | Typical expected density of seawater for this deployment |
| sg_cal_sbect_modes | string | Number of modes to use for thermal-inertia correction | |
| sg_cal_t_g | string | ||
| sg_cal_t_h | string | ||
| sg_cal_t_i | string | ||
| sg_cal_t_j | string | ||
| sg_cal_therm_expan | string | cc/degrees_Celsius | SG thermal expansion coeff |
| sg_cal_volmax | string | m^3 | Maximum displaced volume of the glider |
| sg_cal_wlbb2fl_sig470nm_dark_counts | string | ||
| sg_cal_wlbb2fl_sig470nm_resolution_counts | string | ||
| sg_cal_wlbb2fl_sig470nm_scale_factor | string | ||
| sg_cal_wlbb2fl_sig695nm_dark_counts | string | ||
| sg_cal_wlbb2fl_sig695nm_resolution_counts | string | ||
| sg_cal_wlbb2fl_sig695nm_scale_factor | string | ||
| sg_cal_wlbb2fl_sig700nm_dark_counts | string | ||
| sg_cal_wlbb2fl_sig700nm_resolution_counts | string | ||
| sg_cal_wlbb2fl_sig700nm_scale_factor | string | ||
| start_of_climb_time | string | seconds | Elapsed seconds after dive start when second (positive) apogee pump starts |
| surface_curr_east | string | cm/s | Eastward component of surface current |
| surface_curr_north | string | cm/s | Northward component of surface current |
| surface_curr_qc | string | Whether to trust the surface current values | |
| wlbb2fl | string | ||
| trajectory | trajectory | Dive number for observations |
names = [
'ctd_depth',
'ctd_time',
'ctd_pressure',
'salinity',
'salinity_raw',
'temperature',
'temperature_raw',
'temperature_qc',
'eng_wlbb2fl_sig470nm',
'eng_wlbb2fl_sig695nm',
'eng_wlbb2fl_sig700nm'
]
ds_dict = gt.load.seaglider_basestation_netCDFs(filenames, names, return_merged=True, keep_global_attrs=False)
DIMENSION: sg_data_point
{latitude, longitude, temperature, ctd_pressure, salinity_raw, temperature_qc, ctd_time, salinity, ctd_depth, temperature_raw}
100%|████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 2566/2566 [00:19<00:00, 129.19it/s]
Cannot merge data - not enough time indexed DataFrames
Returning unmerged dataframes
some file lack enough information/details (or are incomplete dives):
early dive issues (2??)
p401067[34]
ds_dict['sg_data_point']
<xarray.Dataset>
Dimensions: (sg_data_point: 349365)
Coordinates:
latitude (sg_data_point) float64 61.48 61.48 61.48 ... 57.93 57.93
longitude (sg_data_point) float64 -173.8 -173.8 ... -169.1 -169.1
ctd_depth (sg_data_point) float64 0.7374 0.2013 ... 1.294 -0.1832
ctd_time_dt64 (sg_data_point) datetime64[ns] 2017-08-13T21:07:40 ... 2...
Dimensions without coordinates: sg_data_point
Data variables:
temperature (sg_data_point) float64 9.512 9.513 9.511 ... 7.004 7.024
ctd_pressure (sg_data_point) float64 0.7451 0.2038 ... 1.306 -0.1844
salinity_raw (sg_data_point) float64 31.16 31.15 31.15 ... 31.46 2.506
temperature_qc (sg_data_point) int64 1 1 1 1 1 1 1 1 1 ... 1 1 1 1 1 1 1 1
ctd_time (sg_data_point) float64 1.503e+09 1.503e+09 ... 1.506e+09
salinity (sg_data_point) float64 nan nan nan nan ... 31.46 nan nan
temperature_raw (sg_data_point) float64 9.512 9.513 9.511 ... 7.004 7.023
dives (sg_data_point) float64 1e+03 1e+03 ... 3.612e+03 3.612e+03
Attributes:
date_created: 2021-07-26 13:55:28
number_of_dives: 3612.0
files: ['p4011000.nc', 'p4011001.nc', 'p4011002.nc', '...
time_coverage_start: 2017-08-13 21:07:40
time_coverage_end: 2017-09-26 16:05:42
geospatial_vertical_min: -0.5030842835078432
geospatial_vertical_max: 67.57316616647842
geospatial_lat_min: 57.89913235353872
geospatial_lat_max: 62.14208166934556
geospatial_lon_min: -174.5055983333333
geospatial_lon_max: -168.8639759711956
processing: [2021-07-26 13:55:28] imported data with Glider...- sg_data_point: 349365
- latitude(sg_data_point)float6461.48 61.48 61.48 ... 57.93 57.93
- comment :
- Latitude of the sample based on hdm DAC
- _FillValue :
- nan
- standard_name :
- latitude
- platform :
- glider
- units :
- degrees_north
- axis :
- Y
array([61.47664333, 61.47664333, 61.47664333, ..., 57.92881377, 57.92883207, 57.92884188]) - longitude(sg_data_point)float64-173.8 -173.8 ... -169.1 -169.1
- comment :
- Longitude of the sample based on hdm DAC
- _FillValue :
- nan
- standard_name :
- longitude
- platform :
- glider
- units :
- degrees_east
- axis :
- X
array([-173.81683 , -173.81683 , -173.81683 , ..., -169.11074467, -169.11078933, -169.11081068]) - ctd_depth(sg_data_point)float640.7374 0.2013 ... 1.294 -0.1832
- comment :
- CTD thermistor depth corrected for average latitude
- platform :
- glider
- standard_name :
- depth
- units :
- meters
- positive :
- down
- axis :
- Z
array([ 0.73742912, 0.20126934, -0.14697653, ..., 2.71163752, 1.29381673, -0.18323594]) - ctd_time_dt64(sg_data_point)datetime64[ns]2017-08-13T21:07:40 ... 2017-09-...
array(['2017-08-13T21:07:40.000000000', '2017-08-13T21:07:54.000000000', '2017-08-13T21:08:02.000000000', ..., '2017-09-26T16:05:18.000000000', '2017-09-26T16:05:30.000000000', '2017-09-26T16:05:42.000000000'], dtype='datetime64[ns]')
- temperature(sg_data_point)float649.512 9.513 9.511 ... 7.004 7.024
- units :
- degrees_Celsius
- comment :
- Termperature (in situ) corrected for thermistor first-order lag
- standard_name :
- sea_water_temperature
- platform :
- glider
array([9.51193002, 9.51302256, 9.51140929, ..., 7.00300615, 7.00403547, 7.02446412]) - ctd_pressure(sg_data_point)float640.7451 0.2038 ... 1.306 -0.1844
- units :
- dbar
- comment :
- Pressure at CTD thermistor
- standard_name :
- sea_water_pressure
- platform :
- glider
array([ 0.74512522, 0.20382167, -0.14781317, ..., 2.73745652, 1.30637229, -0.18443319]) - salinity_raw(sg_data_point)float6431.16 31.15 31.15 ... 31.46 2.506
- units :
- 1e-3
- comment :
- Uncorrected salinity derived from temperature_raw and conductivity_raw (PSU)
- platform :
- glider
array([31.15784827, 31.15432947, 31.15488362, ..., 31.46198175, 31.46214907, 2.50609309]) - temperature_qc(sg_data_point)int641 1 1 1 1 1 1 1 ... 1 1 1 1 1 1 1 1
- flag_meanings :
- QC_NO_CHANGE QC_GOOD QC_PROBABLY_GOOD QC_PROBABLY_BAD QC_BAD QC_CHANGED QC_UNSAMPLED QC_INTERPOLATED QC_MISSING
- comment :
- Whether to trust each corrected temperature value
- flag_values :
- 0 1 2 3 4 5 6 8 9
- platform :
- glider
array([1, 1, 1, ..., 1, 1, 1])
- ctd_time(sg_data_point)float641.503e+09 1.503e+09 ... 1.506e+09
- units :
- seconds since 1970-1-1 00:00:00
- comment :
- Time of CTD sample in GMT epoch format
- standard_name :
- time
- platform :
- glider
- axis :
- T
array([1.50265846e+09, 1.50265847e+09, 1.50265848e+09, ..., 1.50644192e+09, 1.50644193e+09, 1.50644194e+09]) - salinity(sg_data_point)float64nan nan nan nan ... 31.46 nan nan
- units :
- 1e-3
- comment :
- Salinity corrected for thermal-inertia effects (PSU)
- standard_name :
- sea_water_salinity
- platform :
- glider
array([ nan, nan, nan, ..., 31.46201736, nan, nan]) - temperature_raw(sg_data_point)float649.512 9.513 9.511 ... 7.004 7.023
- units :
- degrees_Celsius
- comment :
- Uncorrected temperature (in situ)
- platform :
- glider
array([9.51188082, 9.51303766, 9.51130812, ..., 7.00298831, 7.00352355, 7.02346695]) - dives(sg_data_point)float641e+03 1e+03 ... 3.612e+03 3.612e+03
array([1000. , 1000. , 1000. , ..., 3612.5, 3612.5, 3612.5])
- date_created :
- 2021-07-26 13:55:28
- number_of_dives :
- 3612.0
- files :
- ['p4011000.nc', 'p4011001.nc', 'p4011002.nc', 'p4011003.nc', 'p4011004.nc', 'p4011005.nc', 'p4011006.nc', 'p4011007.nc', 'p4011008.nc', 'p4011009.nc', 'p4011010.nc', 'p4011011.nc', 'p4011012.nc', 'p4011013.nc', 'p4011014.nc', 'p4011015.nc', 'p4011016.nc', 'p4011017.nc', 'p4011018.nc', 'p4011019.nc', 'p4011020.nc', 'p4011021.nc', 'p4011022.nc', 'p4011023.nc', 'p4011024.nc', 'p4011025.nc', 'p4011026.nc', 'p4011027.nc', 'p4011028.nc', 'p4011029.nc', 'p4011030.nc', 'p4011031.nc', 'p4011032.nc', 'p4011033.nc', 'p4011034.nc', 'p4011035.nc', 'p4011036.nc', 'p4011037.nc', 'p4011038.nc', 'p4011039.nc', 'p4011040.nc', 'p4011041.nc', 'p4011042.nc', 'p4011043.nc', 'p4011044.nc', 'p4011045.nc', 'p4011046.nc', 'p4011047.nc', 'p4011048.nc', 'p4011049.nc', 'p4011050.nc', 'p4011051.nc', 'p4011052.nc', 'p4011053.nc', 'p4011054.nc', 'p4011055.nc', 'p4011056.nc', 'p4011057.nc', 'p4011058.nc', 'p4011059.nc', 'p4011060.nc', 'p4011061.nc', 'p4011062.nc', 'p4011063.nc', 'p4011064.nc', 'p4011065.nc', 'p4011066.nc', 'p4011067.nc', 'p4011068.nc', 'p4011069.nc', 'p4011070.nc', 'p4011071.nc', 'p4011072.nc', 'p4011073.nc', 'p4011074.nc', 'p4011075.nc', 'p4011076.nc', 'p4011077.nc', 'p4011078.nc', 'p4011079.nc', 'p4011080.nc', 'p4011081.nc', 'p4011082.nc', 'p4011083.nc', 'p4011084.nc', 'p4011085.nc', 'p4011086.nc', 'p4011087.nc', 'p4011088.nc', 'p4011089.nc', 'p4011090.nc', 'p4011091.nc', 'p4011092.nc', 'p4011093.nc', 'p4011094.nc', 'p4011095.nc', 'p4011096.nc', 'p4011097.nc', 'p4011098.nc', 'p4011099.nc', 'p4011100.nc', 'p4011101.nc', 'p4011102.nc', 'p4011103.nc', 'p4011104.nc', 'p4011105.nc', 'p4011106.nc', 'p4011107.nc', 'p4011108.nc', 'p4011109.nc', 'p4011110.nc', 'p4011111.nc', 'p4011112.nc', 'p4011113.nc', 'p4011114.nc', 'p4011115.nc', 'p4011116.nc', 'p4011117.nc', 'p4011118.nc', 'p4011119.nc', 'p4011120.nc', 'p4011121.nc', 'p4011122.nc', 'p4011123.nc', 'p4011124.nc', 'p4011125.nc', 'p4011126.nc', 'p4011127.nc', 'p4011128.nc', 'p4011129.nc', 'p4011130.nc', 'p4011131.nc', 'p4011132.nc', 'p4011133.nc', 'p4011134.nc', 'p4011135.nc', 'p4011136.nc', 'p4011137.nc', 'p4011138.nc', 'p4011139.nc', 'p4011140.nc', 'p4011141.nc', 'p4011142.nc', 'p4011143.nc', 'p4011144.nc', 'p4011145.nc', 'p4011146.nc', 'p4011147.nc', 'p4011148.nc', 'p4011149.nc', 'p4011150.nc', 'p4011151.nc', 'p4011152.nc', 'p4011153.nc', 'p4011154.nc', 'p4011155.nc', 'p4011156.nc', 'p4011157.nc', 'p4011158.nc', 'p4011159.nc', 'p4011160.nc', 'p4011161.nc', 'p4011162.nc', 'p4011163.nc', 'p4011164.nc', 'p4011165.nc', 'p4011166.nc', 'p4011167.nc', 'p4011168.nc', 'p4011169.nc', 'p4011170.nc', 'p4011171.nc', 'p4011172.nc', 'p4011173.nc', 'p4011174.nc', 'p4011175.nc', 'p4011176.nc', 'p4011177.nc', 'p4011178.nc', 'p4011179.nc', 'p4011180.nc', 'p4011181.nc', 'p4011182.nc', 'p4011183.nc', 'p4011184.nc', 'p4011185.nc', 'p4011186.nc', 'p4011187.nc', 'p4011188.nc', 'p4011189.nc', 'p4011190.nc', 'p4011191.nc', 'p4011192.nc', 'p4011193.nc', 'p4011194.nc', 'p4011195.nc', 'p4011196.nc', 'p4011197.nc', 'p4011198.nc', 'p4011199.nc', 'p4011200.nc', 'p4011201.nc', 'p4011202.nc', 'p4011203.nc', 'p4011204.nc', 'p4011205.nc', 'p4011206.nc', 'p4011207.nc', 'p4011208.nc', 'p4011209.nc', 'p4011210.nc', 'p4011211.nc', 'p4011212.nc', 'p4011213.nc', 'p4011214.nc', 'p4011215.nc', 'p4011216.nc', 'p4011217.nc', 'p4011218.nc', 'p4011219.nc', 'p4011220.nc', 'p4011221.nc', 'p4011222.nc', 'p4011223.nc', 'p4011224.nc', 'p4011225.nc', 'p4011226.nc', 'p4011227.nc', 'p4011228.nc', 'p4011229.nc', 'p4011230.nc', 'p4011231.nc', 'p4011232.nc', 'p4011233.nc', 'p4011234.nc', 'p4011235.nc', 'p4011236.nc', 'p4011237.nc', 'p4011238.nc', 'p4011239.nc', 'p4011240.nc', 'p4011241.nc', 'p4011242.nc', 'p4011243.nc', 'p4011244.nc', 'p4011245.nc', 'p4011246.nc', 'p4011247.nc', 'p4011248.nc', 'p4011249.nc', 'p4011250.nc', 'p4011251.nc', 'p4011252.nc', 'p4011253.nc', 'p4011254.nc', 'p4011255.nc', 'p4011256.nc', 'p4011257.nc', 'p4011258.nc', 'p4011259.nc', 'p4011260.nc', 'p4011261.nc', 'p4011262.nc', 'p4011263.nc', 'p4011264.nc', 'p4011265.nc', 'p4011266.nc', 'p4011267.nc', 'p4011268.nc', 'p4011269.nc', 'p4011270.nc', 'p4011271.nc', 'p4011272.nc', 'p4011273.nc', 'p4011274.nc', 'p4011275.nc', 'p4011276.nc', 'p4011277.nc', 'p4011278.nc', 'p4011279.nc', 'p4011280.nc', 'p4011281.nc', 'p4011282.nc', 'p4011283.nc', 'p4011284.nc', 'p4011285.nc', 'p4011286.nc', 'p4011287.nc', 'p4011288.nc', 'p4011289.nc', 'p4011290.nc', 'p4011291.nc', 'p4011292.nc', 'p4011293.nc', 'p4011294.nc', 'p4011295.nc', 'p4011296.nc', 'p4011297.nc', 'p4011298.nc', 'p4011299.nc', 'p4011300.nc', 'p4011301.nc', 'p4011302.nc', 'p4011303.nc', 'p4011304.nc', 'p4011305.nc', 'p4011306.nc', 'p4011307.nc', 'p4011308.nc', 'p4011309.nc', 'p4011310.nc', 'p4011311.nc', 'p4011312.nc', 'p4011313.nc', 'p4011314.nc', 'p4011315.nc', 'p4011316.nc', 'p4011317.nc', 'p4011318.nc', 'p4011319.nc', 'p4011320.nc', 'p4011321.nc', 'p4011322.nc', 'p4011323.nc', 'p4011324.nc', 'p4011325.nc', 'p4011326.nc', 'p4011327.nc', 'p4011328.nc', 'p4011329.nc', 'p4011330.nc', 'p4011331.nc', 'p4011332.nc', 'p4011333.nc', 'p4011334.nc', 'p4011335.nc', 'p4011336.nc', 'p4011337.nc', 'p4011338.nc', 'p4011339.nc', 'p4011340.nc', 'p4011341.nc', 'p4011342.nc', 'p4011343.nc', 'p4011344.nc', 'p4011345.nc', 'p4011346.nc', 'p4011347.nc', 'p4011348.nc', 'p4011349.nc', 'p4011350.nc', 'p4011351.nc', 'p4011352.nc', 'p4011353.nc', 'p4011354.nc', 'p4011355.nc', 'p4011356.nc', 'p4011357.nc', 'p4011358.nc', 'p4011359.nc', 'p4011360.nc', 'p4011361.nc', 'p4011362.nc', 'p4011363.nc', 'p4011364.nc', 'p4011365.nc', 'p4011366.nc', 'p4011367.nc', 'p4011368.nc', 'p4011369.nc', 'p4011370.nc', 'p4011371.nc', 'p4011372.nc', 'p4011373.nc', 'p4011374.nc', 'p4011375.nc', 'p4011376.nc', 'p4011377.nc', 'p4011378.nc', 'p4011379.nc', 'p4011380.nc', 'p4011381.nc', 'p4011382.nc', 'p4011383.nc', 'p4011384.nc', 'p4011385.nc', 'p4011386.nc', 'p4011387.nc', 'p4011388.nc', 'p4011389.nc', 'p4011390.nc', 'p4011391.nc', 'p4011392.nc', 'p4011393.nc', 'p4011394.nc', 'p4011395.nc', 'p4011396.nc', 'p4011397.nc', 'p4011398.nc', 'p4011399.nc', 'p4011400.nc', 'p4011401.nc', 'p4011402.nc', 'p4011403.nc', 'p4011404.nc', 'p4011405.nc', 'p4011406.nc', 'p4011407.nc', 'p4011408.nc', 'p4011409.nc', 'p4011410.nc', 'p4011411.nc', 'p4011412.nc', 'p4011413.nc', 'p4011414.nc', 'p4011415.nc', 'p4011416.nc', 'p4011417.nc', 'p4011418.nc', 'p4011419.nc', 'p4011420.nc', 'p4011421.nc', 'p4011422.nc', 'p4011423.nc', 'p4011424.nc', 'p4011425.nc', 'p4011426.nc', 'p4011427.nc', 'p4011428.nc', 'p4011429.nc', 'p4011430.nc', 'p4011431.nc', 'p4011432.nc', 'p4011433.nc', 'p4011434.nc', 'p4011435.nc', 'p4011436.nc', 'p4011437.nc', 'p4011438.nc', 'p4011439.nc', 'p4011440.nc', 'p4011441.nc', 'p4011442.nc', 'p4011443.nc', 'p4011444.nc', 'p4011445.nc', 'p4011446.nc', 'p4011447.nc', 'p4011448.nc', 'p4011449.nc', 'p4011450.nc', 'p4011451.nc', 'p4011452.nc', 'p4011453.nc', 'p4011454.nc', 'p4011455.nc', 'p4011456.nc', 'p4011457.nc', 'p4011458.nc', 'p4011459.nc', 'p4011460.nc', 'p4011461.nc', 'p4011462.nc', 'p4011463.nc', 'p4011464.nc', 'p4011465.nc', 'p4011466.nc', 'p4011467.nc', 'p4011468.nc', 'p4011469.nc', 'p4011470.nc', 'p4011471.nc', 'p4011472.nc', 'p4011473.nc', 'p4011474.nc', 'p4011475.nc', 'p4011476.nc', 'p4011477.nc', 'p4011478.nc', 'p4011479.nc', 'p4011480.nc', 'p4011481.nc', 'p4011482.nc', 'p4011483.nc', 'p4011484.nc', 'p4011485.nc', 'p4011486.nc', 'p4011487.nc', 'p4011488.nc', 'p4011489.nc', 'p4011490.nc', 'p4011491.nc', 'p4011492.nc', 'p4011493.nc', 'p4011494.nc', 'p4011495.nc', 'p4011496.nc', 'p4011497.nc', 'p4011498.nc', 'p4011499.nc', 'p4011500.nc', 'p4011501.nc', 'p4011502.nc', 'p4011503.nc', 'p4011504.nc', 'p4011505.nc', 'p4011506.nc', 'p4011507.nc', 'p4011508.nc', 'p4011509.nc', 'p4011510.nc', 'p4011511.nc', 'p4011512.nc', 'p4011513.nc', 'p4011514.nc', 'p4011515.nc', 'p4011516.nc', 'p4011517.nc', 'p4011518.nc', 'p4011519.nc', 'p4011520.nc', 'p4011521.nc', 'p4011522.nc', 'p4011523.nc', 'p4011524.nc', 'p4011525.nc', 'p4011526.nc', 'p4011527.nc', 'p4011528.nc', 'p4011529.nc', 'p4011530.nc', 'p4011531.nc', 'p4011532.nc', 'p4011533.nc', 'p4011534.nc', 'p4011535.nc', 'p4011536.nc', 'p4011537.nc', 'p4011538.nc', 'p4011539.nc', 'p4011540.nc', 'p4011541.nc', 'p4011542.nc', 'p4011543.nc', 'p4011544.nc', 'p4011545.nc', 'p4011546.nc', 'p4011547.nc', 'p4011548.nc', 'p4011549.nc', 'p4011550.nc', 'p4011551.nc', 'p4011552.nc', 'p4011553.nc', 'p4011554.nc', 'p4011555.nc', 'p4011556.nc', 'p4011557.nc', 'p4011558.nc', 'p4011559.nc', 'p4011560.nc', 'p4011561.nc', 'p4011562.nc', 'p4011563.nc', 'p4011564.nc', 'p4011565.nc', 'p4011566.nc', 'p4011567.nc', 'p4011568.nc', 'p4011569.nc', 'p4011570.nc', 'p4011571.nc', 'p4011572.nc', 'p4011573.nc', 'p4011574.nc', 'p4011575.nc', 'p4011576.nc', 'p4011577.nc', 'p4011578.nc', 'p4011579.nc', 'p4011580.nc', 'p4011581.nc', 'p4011582.nc', 'p4011583.nc', 'p4011584.nc', 'p4011585.nc', 'p4011586.nc', 'p4011587.nc', 'p4011588.nc', 'p4011589.nc', 'p4011590.nc', 'p4011591.nc', 'p4011592.nc', 'p4011593.nc', 'p4011594.nc', 'p4011595.nc', 'p4011596.nc', 'p4011597.nc', 'p4011598.nc', 'p4011599.nc', 'p4011600.nc', 'p4011601.nc', 'p4011602.nc', 'p4011603.nc', 'p4011604.nc', 'p4011605.nc', 'p4011606.nc', 'p4011607.nc', 'p4011608.nc', 'p4011609.nc', 'p4011610.nc', 'p4011611.nc', 'p4011612.nc', 'p4011613.nc', 'p4011614.nc', 'p4011615.nc', 'p4011616.nc', 'p4011617.nc', 'p4011618.nc', 'p4011619.nc', 'p4011620.nc', 'p4011621.nc', 'p4011622.nc', 'p4011623.nc', 'p4011624.nc', 'p4011625.nc', 'p4011626.nc', 'p4011627.nc', 'p4011628.nc', 'p4011629.nc', 'p4011630.nc', 'p4011631.nc', 'p4011632.nc', 'p4011633.nc', 'p4011634.nc', 'p4011635.nc', 'p4011636.nc', 'p4011637.nc', 'p4011638.nc', 'p4011639.nc', 'p4011640.nc', 'p4011641.nc', 'p4011642.nc', 'p4011643.nc', 'p4011644.nc', 'p4011645.nc', 'p4011646.nc', 'p4011647.nc', 'p4011648.nc', 'p4011649.nc', 'p4011650.nc', 'p4011651.nc', 'p4011652.nc', 'p4011653.nc', 'p4011654.nc', 'p4011655.nc', 'p4011656.nc', 'p4011657.nc', 'p4011658.nc', 'p4011659.nc', 'p4011660.nc', 'p4011661.nc', 'p4011662.nc', 'p4011663.nc', 'p4011664.nc', 'p4011665.nc', 'p4011666.nc', 'p4011667.nc', 'p4011668.nc', 'p4011669.nc', 'p4011670.nc', 'p4011671.nc', 'p4011672.nc', 'p4011673.nc', 'p4011674.nc', 'p4011675.nc', 'p4011676.nc', 'p4011677.nc', 'p4011678.nc', 'p4011679.nc', 'p4011680.nc', 'p4011681.nc', 'p4011682.nc', 'p4011683.nc', 'p4011684.nc', 'p4011685.nc', 'p4011686.nc', 'p4011687.nc', 'p4011688.nc', 'p4011689.nc', 'p4011690.nc', 'p4011691.nc', 'p4011692.nc', 'p4011693.nc', 'p4011694.nc', 'p4011695.nc', 'p4011696.nc', 'p4011697.nc', 'p4011698.nc', 'p4011699.nc', 'p4011700.nc', 'p4011701.nc', 'p4011702.nc', 'p4011703.nc', 'p4011704.nc', 'p4011705.nc', 'p4011706.nc', 'p4011707.nc', 'p4011708.nc', 'p4011709.nc', 'p4011710.nc', 'p4011711.nc', 'p4011712.nc', 'p4011713.nc', 'p4011714.nc', 'p4011715.nc', 'p4011716.nc', 'p4011717.nc', 'p4011718.nc', 'p4011719.nc', 'p4011720.nc', 'p4011721.nc', 'p4011722.nc', 'p4011723.nc', 'p4011724.nc', 'p4011725.nc', 'p4011726.nc', 'p4011727.nc', 'p4011728.nc', 'p4011729.nc', 'p4011730.nc', 'p4011731.nc', 'p4011732.nc', 'p4011733.nc', 'p4011734.nc', 'p4011735.nc', 'p4011736.nc', 'p4011737.nc', 'p4011738.nc', 'p4011739.nc', 'p4011740.nc', 'p4011741.nc', 'p4011742.nc', 'p4011743.nc', 'p4011744.nc', 'p4011745.nc', 'p4011746.nc', 'p4011747.nc', 'p4011748.nc', 'p4011749.nc', 'p4011750.nc', 'p4011751.nc', 'p4011752.nc', 'p4011753.nc', 'p4011754.nc', 'p4011755.nc', 'p4011756.nc', 'p4011757.nc', 'p4011758.nc', 'p4011759.nc', 'p4011760.nc', 'p4011761.nc', 'p4011762.nc', 'p4011763.nc', 'p4011764.nc', 'p4011765.nc', 'p4011766.nc', 'p4011767.nc', 'p4011768.nc', 'p4011769.nc', 'p4011770.nc', 'p4011771.nc', 'p4011772.nc', 'p4011773.nc', 'p4011774.nc', 'p4011775.nc', 'p4011776.nc', 'p4011777.nc', 'p4011778.nc', 'p4011779.nc', 'p4011780.nc', 'p4011781.nc', 'p4011782.nc', 'p4011783.nc', 'p4011784.nc', 'p4011785.nc', 'p4011786.nc', 'p4011787.nc', 'p4011788.nc', 'p4011789.nc', 'p4011790.nc', 'p4011791.nc', 'p4011792.nc', 'p4011793.nc', 'p4011794.nc', 'p4011795.nc', 'p4011796.nc', 'p4011797.nc', 'p4011798.nc', 'p4011799.nc', 'p4011800.nc', 'p4011801.nc', 'p4011802.nc', 'p4011803.nc', 'p4011804.nc', 'p4011805.nc', 'p4011806.nc', 'p4011807.nc', 'p4011808.nc', 'p4011809.nc', 'p4011810.nc', 'p4011811.nc', 'p4011812.nc', 'p4011813.nc', 'p4011814.nc', 'p4011815.nc', 'p4011816.nc', 'p4011817.nc', 'p4011818.nc', 'p4011819.nc', 'p4011820.nc', 'p4011821.nc', 'p4011822.nc', 'p4011823.nc', 'p4011824.nc', 'p4011825.nc', 'p4011826.nc', 'p4011827.nc', 'p4011828.nc', 'p4011829.nc', 'p4011830.nc', 'p4011831.nc', 'p4011832.nc', 'p4011833.nc', 'p4011834.nc', 'p4011835.nc', 'p4011836.nc', 'p4011837.nc', 'p4011838.nc', 'p4011839.nc', 'p4011840.nc', 'p4011841.nc', 'p4011842.nc', 'p4011843.nc', 'p4011844.nc', 'p4011845.nc', 'p4011846.nc', 'p4011847.nc', 'p4011848.nc', 'p4011849.nc', 'p4011850.nc', 'p4011851.nc', 'p4011852.nc', 'p4011853.nc', 'p4011854.nc', 'p4011855.nc', 'p4011856.nc', 'p4011857.nc', 'p4011858.nc', 'p4011859.nc', 'p4011860.nc', 'p4011861.nc', 'p4011862.nc', 'p4011863.nc', 'p4011864.nc', 'p4011865.nc', 'p4011866.nc', 'p4011867.nc', 'p4011868.nc', 'p4011869.nc', 'p4011870.nc', 'p4011871.nc', 'p4011872.nc', 'p4011873.nc', 'p4011874.nc', 'p4011875.nc', 'p4011876.nc', 'p4011877.nc', 'p4011878.nc', 'p4011879.nc', 'p4011880.nc', 'p4011881.nc', 'p4011882.nc', 'p4011883.nc', 'p4011884.nc', 'p4011885.nc', 'p4011886.nc', 'p4011887.nc', 'p4011888.nc', 'p4011889.nc', 'p4011890.nc', 'p4011891.nc', 'p4011892.nc', 'p4011893.nc', 'p4011894.nc', 'p4011895.nc', 'p4011896.nc', 'p4011897.nc', 'p4011898.nc', 'p4011899.nc', 'p4011900.nc', 'p4011901.nc', 'p4011902.nc', 'p4011903.nc', 'p4011904.nc', 'p4011905.nc', 'p4011906.nc', 'p4011907.nc', 'p4011908.nc', 'p4011909.nc', 'p4011910.nc', 'p4011911.nc', 'p4011912.nc', 'p4011913.nc', 'p4011914.nc', 'p4011915.nc', 'p4011916.nc', 'p4011917.nc', 'p4011918.nc', 'p4011919.nc', 'p4011920.nc', 'p4011921.nc', 'p4011922.nc', 'p4011923.nc', 'p4011924.nc', 'p4011925.nc', 'p4011926.nc', 'p4011927.nc', 'p4011928.nc', 'p4011929.nc', 'p4011930.nc', 'p4011931.nc', 'p4011932.nc', 'p4011933.nc', 'p4011934.nc', 'p4011935.nc', 'p4011936.nc', 'p4011937.nc', 'p4011938.nc', 'p4011939.nc', 'p4011940.nc', 'p4011941.nc', 'p4011942.nc', 'p4011943.nc', 'p4011944.nc', 'p4011945.nc', 'p4011946.nc', 'p4011947.nc', 'p4011948.nc', 'p4011949.nc', 'p4011950.nc', 'p4011951.nc', 'p4011952.nc', 'p4011953.nc', 'p4011954.nc', 'p4011955.nc', 'p4011956.nc', 'p4011957.nc', 'p4011958.nc', 'p4011959.nc', 'p4011960.nc', 'p4011961.nc', 'p4011962.nc', 'p4011963.nc', 'p4011964.nc', 'p4011965.nc', 'p4011966.nc', 'p4011967.nc', 'p4011968.nc', 'p4011969.nc', 'p4011970.nc', 'p4011971.nc', 'p4011972.nc', 'p4011973.nc', 'p4011974.nc', 'p4011975.nc', 'p4011976.nc', 'p4011977.nc', 'p4011978.nc', 'p4011979.nc', 'p4011980.nc', 'p4011981.nc', 'p4011982.nc', 'p4011983.nc', 'p4011984.nc', 'p4011985.nc', 'p4011986.nc', 'p4011987.nc', 'p4011988.nc', 'p4011989.nc', 'p4011990.nc', 'p4011991.nc', 'p4011992.nc', 'p4011993.nc', 'p4011994.nc', 'p4011995.nc', 'p4011996.nc', 'p4011997.nc', 'p4011998.nc', 'p4011999.nc', 'p4012000.nc', 'p4012001.nc', 'p4012002.nc', 'p4012003.nc', 'p4012004.nc', 'p4012005.nc', 'p4012006.nc', 'p4012007.nc', 'p4012008.nc', 'p4012009.nc', 'p4012010.nc', 'p4012011.nc', 'p4012012.nc', 'p4012013.nc', 'p4012014.nc', 'p4012015.nc', 'p4012016.nc', 'p4012017.nc', 'p4012018.nc', 'p4012019.nc', 'p4012020.nc', 'p4012021.nc', 'p4012022.nc', 'p4012023.nc', 'p4012024.nc', 'p4012025.nc', 'p4012026.nc', 'p4012027.nc', 'p4012028.nc', 'p4012029.nc', 'p4012030.nc', 'p4012031.nc', 'p4012032.nc', 'p4012033.nc', 'p4012034.nc', 'p4012035.nc', 'p4012036.nc', 'p4012037.nc', 'p4012038.nc', 'p4012039.nc', 'p4012040.nc', 'p4012041.nc', 'p4012042.nc', 'p4012043.nc', 'p4012044.nc', 'p4012045.nc', 'p4012046.nc', 'p4012047.nc', 'p4012048.nc', 'p4012049.nc', 'p4012050.nc', 'p4012051.nc', 'p4012052.nc', 'p4012053.nc', 'p4012054.nc', 'p4012055.nc', 'p4012056.nc', 'p4012057.nc', 'p4012058.nc', 'p4012059.nc', 'p4012060.nc', 'p4012061.nc', 'p4012062.nc', 'p4012063.nc', 'p4012064.nc', 'p4012065.nc', 'p4012066.nc', 'p4012067.nc', 'p4012068.nc', 'p4012069.nc', 'p4012070.nc', 'p4012071.nc', 'p4012072.nc', 'p4012073.nc', 'p4012074.nc', 'p4012075.nc', 'p4012076.nc', 'p4012077.nc', 'p4012078.nc', 'p4012079.nc', 'p4012080.nc', 'p4012081.nc', 'p4012082.nc', 'p4012083.nc', 'p4012084.nc', 'p4012085.nc', 'p4012086.nc', 'p4012087.nc', 'p4012088.nc', 'p4012089.nc', 'p4012090.nc', 'p4012091.nc', 'p4012092.nc', 'p4012093.nc', 'p4012094.nc', 'p4012095.nc', 'p4012096.nc', 'p4012097.nc', 'p4012098.nc', 'p4012099.nc', 'p4012100.nc', 'p4012101.nc', 'p4012102.nc', 'p4012103.nc', 'p4012104.nc', 'p4012105.nc', 'p4012106.nc', 'p4012107.nc', 'p4012108.nc', 'p4012109.nc', 'p4012110.nc', 'p4012111.nc', 'p4012112.nc', 'p4012113.nc', 'p4012114.nc', 'p4012115.nc', 'p4012116.nc', 'p4012117.nc', 'p4012118.nc', 'p4012119.nc', 'p4012120.nc', 'p4012121.nc', 'p4012122.nc', 'p4012123.nc', 'p4012124.nc', 'p4012125.nc', 'p4012126.nc', 'p4012127.nc', 'p4012128.nc', 'p4012129.nc', 'p4012130.nc', 'p4012131.nc', 'p4012132.nc', 'p4012133.nc', 'p4012134.nc', 'p4012135.nc', 'p4012136.nc', 'p4012137.nc', 'p4012138.nc', 'p4012139.nc', 'p4012140.nc', 'p4012141.nc', 'p4012142.nc', 'p4012143.nc', 'p4012144.nc', 'p4012145.nc', 'p4012146.nc', 'p4012147.nc', 'p4012148.nc', 'p4012149.nc', 'p4012150.nc', 'p4012151.nc', 'p4012152.nc', 'p4012153.nc', 'p4012154.nc', 'p4012155.nc', 'p4012156.nc', 'p4012157.nc', 'p4012158.nc', 'p4012159.nc', 'p4012160.nc', 'p4012161.nc', 'p4012162.nc', 'p4012163.nc', 'p4012164.nc', 'p4012165.nc', 'p4012166.nc', 'p4012167.nc', 'p4012168.nc', 'p4012169.nc', 'p4012170.nc', 'p4012171.nc', 'p4012172.nc', 'p4012173.nc', 'p4012174.nc', 'p4012175.nc', 'p4012176.nc', 'p4012177.nc', 'p4012178.nc', 'p4012179.nc', 'p4012180.nc', 'p4012181.nc', 'p4012182.nc', 'p4012183.nc', 'p4012184.nc', 'p4012185.nc', 'p4012186.nc', 'p4012187.nc', 'p4012188.nc', 'p4012189.nc', 'p4012190.nc', 'p4012191.nc', 'p4012192.nc', 'p4012193.nc', 'p4012194.nc', 'p4012195.nc', 'p4012196.nc', 'p4012197.nc', 'p4012198.nc', 'p4012199.nc', 'p4012200.nc', 'p4012201.nc', 'p4012202.nc', 'p4012203.nc', 'p4012204.nc', 'p4012205.nc', 'p4012206.nc', 'p4012207.nc', 'p4012208.nc', 'p4012209.nc', 'p4012210.nc', 'p4012211.nc', 'p4012212.nc', 'p4012213.nc', 'p4012214.nc', 'p4012215.nc', 'p4012216.nc', 'p4012217.nc', 'p4012218.nc', 'p4012219.nc', 'p4012220.nc', 'p4012221.nc', 'p4012222.nc', 'p4012223.nc', 'p4012224.nc', 'p4012225.nc', 'p4012226.nc', 'p4012227.nc', 'p4012228.nc', 'p4012229.nc', 'p4012230.nc', 'p4012231.nc', 'p4012232.nc', 'p4012233.nc', 'p4012234.nc', 'p4012235.nc', 'p4012236.nc', 'p4012237.nc', 'p4012238.nc', 'p4012239.nc', 'p4012240.nc', 'p4012241.nc', 'p4012242.nc', 'p4012243.nc', 'p4012244.nc', 'p4012245.nc', 'p4012246.nc', 'p4012247.nc', 'p4012248.nc', 'p4012249.nc', 'p4012250.nc', 'p4012251.nc', 'p4012252.nc', 'p4012253.nc', 'p4012254.nc', 'p4012255.nc', 'p4012256.nc', 'p4012257.nc', 'p4012258.nc', 'p4012259.nc', 'p4012260.nc', 'p4012261.nc', 'p4012262.nc', 'p4012263.nc', 'p4012264.nc', 'p4012265.nc', 'p4012266.nc', 'p4012267.nc', 'p4012268.nc', 'p4012269.nc', 'p4012270.nc', 'p4012271.nc', 'p4012272.nc', 'p4012273.nc', 'p4012274.nc', 'p4012275.nc', 'p4012276.nc', 'p4012277.nc', 'p4012278.nc', 'p4012279.nc', 'p4012280.nc', 'p4012281.nc', 'p4012282.nc', 'p4012283.nc', 'p4012284.nc', 'p4012285.nc', 'p4012286.nc', 'p4012287.nc', 'p4012288.nc', 'p4012289.nc', 'p4012290.nc', 'p4012291.nc', 'p4012292.nc', 'p4012293.nc', 'p4012294.nc', 'p4012295.nc', 'p4012296.nc', 'p4012297.nc', 'p4012298.nc', 'p4012299.nc', 'p4012300.nc', 'p4012301.nc', 'p4012302.nc', 'p4012303.nc', 'p4012304.nc', 'p4012305.nc', 'p4012306.nc', 'p4012307.nc', 'p4012308.nc', 'p4012309.nc', 'p4012310.nc', 'p4012311.nc', 'p4012312.nc', 'p4012313.nc', 'p4012314.nc', 'p4012315.nc', 'p4012316.nc', 'p4012317.nc', 'p4012318.nc', 'p4012319.nc', 'p4012320.nc', 'p4012321.nc', 'p4012322.nc', 'p4012323.nc', 'p4012324.nc', 'p4012325.nc', 'p4012326.nc', 'p4012327.nc', 'p4012328.nc', 'p4012329.nc', 'p4012330.nc', 'p4012331.nc', 'p4012332.nc', 'p4012333.nc', 'p4012334.nc', 'p4012335.nc', 'p4012336.nc', 'p4012337.nc', 'p4012338.nc', 'p4012339.nc', 'p4012340.nc', 'p4012341.nc', 'p4012342.nc', 'p4012343.nc', 'p4012344.nc', 'p4012345.nc', 'p4012346.nc', 'p4012347.nc', 'p4012348.nc', 'p4012349.nc', 'p4012350.nc', 'p4012351.nc', 'p4012352.nc', 'p4012353.nc', 'p4012354.nc', 'p4012355.nc', 'p4012356.nc', 'p4012357.nc', 'p4012358.nc', 'p4012359.nc', 'p4012360.nc', 'p4012361.nc', 'p4012362.nc', 'p4012363.nc', 'p4012364.nc', 'p4012365.nc', 'p4012366.nc', 'p4012367.nc', 'p4012368.nc', 'p4012369.nc', 'p4012370.nc', 'p4012371.nc', 'p4012372.nc', 'p4012373.nc', 'p4012374.nc', 'p4012375.nc', 'p4012376.nc', 'p4012377.nc', 'p4012378.nc', 'p4012379.nc', 'p4012380.nc', 'p4012381.nc', 'p4012382.nc', 'p4012383.nc', 'p4012384.nc', 'p4012385.nc', 'p4012386.nc', 'p4012387.nc', 'p4012388.nc', 'p4012389.nc', 'p4012390.nc', 'p4012391.nc', 'p4012392.nc', 'p4012393.nc', 'p4012394.nc', 'p4012395.nc', 'p4012396.nc', 'p4012397.nc', 'p4012398.nc', 'p4012399.nc', 'p4012400.nc', 'p4012401.nc', 'p4012402.nc', 'p4012403.nc', 'p4012404.nc', 'p4012405.nc', 'p4012406.nc', 'p4012407.nc', 'p4012408.nc', 'p4012409.nc', 'p4012410.nc', 'p4012411.nc', 'p4012412.nc', 'p4012413.nc', 'p4012414.nc', 'p4012415.nc', 'p4012416.nc', 'p4012417.nc', 'p4012418.nc', 'p4012419.nc', 'p4012420.nc', 'p4012421.nc', 'p4012422.nc', 'p4012423.nc', 'p4012424.nc', 'p4012425.nc', 'p4012426.nc', 'p4012427.nc', 'p4012428.nc', 'p4012429.nc', 'p4012430.nc', 'p4012431.nc', 'p4012432.nc', 'p4012433.nc', 'p4012434.nc', 'p4012435.nc', 'p4012436.nc', 'p4012437.nc', 'p4012438.nc', 'p4012439.nc', 'p4012440.nc', 'p4012441.nc', 'p4012442.nc', 'p4012443.nc', 'p4012444.nc', 'p4012445.nc', 'p4012446.nc', 'p4012447.nc', 'p4012448.nc', 'p4012449.nc', 'p4012450.nc', 'p4012451.nc', 'p4012452.nc', 'p4012453.nc', 'p4012454.nc', 'p4012455.nc', 'p4012456.nc', 'p4012457.nc', 'p4012458.nc', 'p4012459.nc', 'p4012460.nc', 'p4012461.nc', 'p4012462.nc', 'p4012463.nc', 'p4012464.nc', 'p4012465.nc', 'p4012466.nc', 'p4012467.nc', 'p4012468.nc', 'p4012469.nc', 'p4012470.nc', 'p4012471.nc', 'p4012472.nc', 'p4012473.nc', 'p4012474.nc', 'p4012475.nc', 'p4012476.nc', 'p4012477.nc', 'p4012478.nc', 'p4012479.nc', 'p4012480.nc', 'p4012481.nc', 'p4012482.nc', 'p4012483.nc', 'p4012484.nc', 'p4012485.nc', 'p4012486.nc', 'p4012487.nc', 'p4012488.nc', 'p4012489.nc', 'p4012490.nc', 'p4012491.nc', 'p4012492.nc', 'p4012493.nc', 'p4012494.nc', 'p4012495.nc', 'p4012496.nc', 'p4012497.nc', 'p4012498.nc', 'p4012499.nc', 'p4012500.nc', 'p4012501.nc', 'p4012502.nc', 'p4012503.nc', 'p4012504.nc', 'p4012505.nc', 'p4012506.nc', 'p4012507.nc', 'p4012508.nc', 'p4012509.nc', 'p4012510.nc', 'p4012511.nc', 'p4012512.nc', 'p4012513.nc', 'p4012514.nc', 'p4012515.nc', 'p4012516.nc', 'p4012517.nc', 'p4012518.nc', 'p4012519.nc', 'p4012520.nc', 'p4012521.nc', 'p4012522.nc', 'p4012523.nc', 'p4012524.nc', 'p4012525.nc', 'p4012526.nc', 'p4012527.nc', 'p4012528.nc', 'p4012529.nc', 'p4012530.nc', 'p4012531.nc', 'p4012532.nc', 'p4012533.nc', 'p4012534.nc', 'p4012535.nc', 'p4012536.nc', 'p4012537.nc', 'p4012538.nc', 'p4012539.nc', 'p4012540.nc', 'p4012541.nc', 'p4012542.nc', 'p4012543.nc', 'p4012544.nc', 'p4012545.nc', 'p4012546.nc', 'p4012547.nc', 'p4012548.nc', 'p4012549.nc', 'p4012550.nc', 'p4012551.nc', 'p4012552.nc', 'p4012553.nc', 'p4012554.nc', 'p4012555.nc', 'p4012556.nc', 'p4012557.nc', 'p4012558.nc', 'p4012559.nc', 'p4012560.nc', 'p4012561.nc', 'p4012562.nc', 'p4012563.nc', 'p4012564.nc', 'p4012565.nc', 'p4012566.nc', 'p4012567.nc', 'p4012568.nc', 'p4012569.nc', 'p4012570.nc', 'p4012571.nc', 'p4012572.nc', 'p4012573.nc', 'p4012574.nc', 'p4012575.nc', 'p4012576.nc', 'p4012577.nc', 'p4012578.nc', 'p4012579.nc', 'p4012580.nc', 'p4012581.nc', 'p4012582.nc', 'p4012583.nc', 'p4012584.nc', 'p4012585.nc', 'p4012586.nc', 'p4012587.nc', 'p4012588.nc', 'p4012589.nc', 'p4012590.nc', 'p4012591.nc', 'p4012592.nc', 'p4012593.nc', 'p4012594.nc', 'p4012595.nc', 'p4012596.nc', 'p4012597.nc', 'p4012598.nc', 'p4012599.nc', 'p4012600.nc', 'p4012601.nc', 'p4012602.nc', 'p4012603.nc', 'p4012604.nc', 'p4012605.nc', 'p4012606.nc', 'p4012607.nc', 'p4012608.nc', 'p4012609.nc', 'p4012610.nc', 'p4012611.nc', 'p4012612.nc', 'p4012613.nc', 'p4012614.nc', 'p4012615.nc', 'p4012616.nc', 'p4012617.nc', 'p4012618.nc', 'p4012619.nc', 'p4012620.nc', 'p4012621.nc', 'p4012622.nc', 'p4012623.nc', 'p4012624.nc', 'p4012625.nc', 'p4012626.nc', 'p4012627.nc', 'p4012628.nc', 'p4012629.nc', 'p4012630.nc', 'p4012631.nc', 'p4012632.nc', 'p4012633.nc', 'p4012634.nc', 'p4012635.nc', 'p4012636.nc', 'p4012637.nc', 'p4012638.nc', 'p4012639.nc', 'p4012640.nc', 'p4012641.nc', 'p4012642.nc', 'p4012643.nc', 'p4012644.nc', 'p4012645.nc', 'p4012646.nc', 'p4012647.nc', 'p4012648.nc', 'p4012649.nc', 'p4012650.nc', 'p4012651.nc', 'p4012652.nc', 'p4012653.nc', 'p4012654.nc', 'p4012655.nc', 'p4012656.nc', 'p4012657.nc', 'p4012658.nc', 'p4012659.nc', 'p4012660.nc', 'p4012661.nc', 'p4012662.nc', 'p4012663.nc', 'p4012664.nc', 'p4012665.nc', 'p4012666.nc', 'p4012667.nc', 'p4012668.nc', 'p4012669.nc', 'p4012670.nc', 'p4012671.nc', 'p4012672.nc', 'p4012673.nc', 'p4012674.nc', 'p4012675.nc', 'p4012676.nc', 'p4012677.nc', 'p4012678.nc', 'p4012679.nc', 'p4012680.nc', 'p4012681.nc', 'p4012682.nc', 'p4012683.nc', 'p4012684.nc', 'p4012685.nc', 'p4012686.nc', 'p4012687.nc', 'p4012688.nc', 'p4012689.nc', 'p4012690.nc', 'p4012691.nc', 'p4012692.nc', 'p4012693.nc', 'p4012694.nc', 'p4012695.nc', 'p4012696.nc', 'p4012697.nc', 'p4012698.nc', 'p4012699.nc', 'p4012700.nc', 'p4012701.nc', 'p4012702.nc', 'p4012703.nc', 'p4012704.nc', 'p4012705.nc', 'p4012706.nc', 'p4012707.nc', 'p4012708.nc', 'p4012709.nc', 'p4012710.nc', 'p4012711.nc', 'p4012712.nc', 'p4012713.nc', 'p4012714.nc', 'p4012715.nc', 'p4012717.nc', 'p4012762.nc', 'p4012763.nc', 'p4012764.nc', 'p4012765.nc', 'p4012766.nc', 'p4012767.nc', 'p4012768.nc', 'p4012769.nc', 'p4012770.nc', 'p4012771.nc', 'p4012772.nc', 'p4012773.nc', 'p4012774.nc', 'p4012775.nc', 'p4012776.nc', 'p4012777.nc', 'p4012778.nc', 'p4012779.nc', 'p4012780.nc', 'p4012781.nc', 'p4012782.nc', 'p4012783.nc', 'p4012784.nc', 'p4012785.nc', 'p4012786.nc', 'p4012787.nc', 'p4012788.nc', 'p4012789.nc', 'p4012790.nc', 'p4012791.nc', 'p4012792.nc', 'p4012793.nc', 'p4012794.nc', 'p4012795.nc', 'p4012796.nc', 'p4012797.nc', 'p4012798.nc', 'p4012799.nc', 'p4012800.nc', 'p4012801.nc', 'p4012802.nc', 'p4012803.nc', 'p4012804.nc', 'p4012805.nc', 'p4012806.nc', 'p4012807.nc', 'p4012808.nc', 'p4012809.nc', 'p4012810.nc', 'p4012811.nc', 'p4012812.nc', 'p4012813.nc', 'p4012814.nc', 'p4012815.nc', 'p4012816.nc', 'p4012817.nc', 'p4012818.nc', 'p4012819.nc', 'p4012820.nc', 'p4012821.nc', 'p4012822.nc', 'p4012823.nc', 'p4012824.nc', 'p4012825.nc', 'p4012826.nc', 'p4012827.nc', 'p4012828.nc', 'p4012829.nc', 'p4012830.nc', 'p4012831.nc', 'p4012832.nc', 'p4012833.nc', 'p4012834.nc', 'p4012835.nc', 'p4012836.nc', 'p4012837.nc', 'p4012838.nc', 'p4012839.nc', 'p4012840.nc', 'p4012841.nc', 'p4012842.nc', 'p4012843.nc', 'p4012844.nc', 'p4012845.nc', 'p4012846.nc', 'p4012847.nc', 'p4012848.nc', 'p4012849.nc', 'p4012850.nc', 'p4012851.nc', 'p4012852.nc', 'p4012853.nc', 'p4012854.nc', 'p4012855.nc', 'p4012856.nc', 'p4012857.nc', 'p4012858.nc', 'p4012859.nc', 'p4012860.nc', 'p4012861.nc', 'p4012862.nc', 'p4012863.nc', 'p4012864.nc', 'p4012865.nc', 'p4012866.nc', 'p4012867.nc', 'p4012868.nc', 'p4012869.nc', 'p4012870.nc', 'p4012871.nc', 'p4012872.nc', 'p4012873.nc', 'p4012874.nc', 'p4012875.nc', 'p4012876.nc', 'p4012877.nc', 'p4012878.nc', 'p4012879.nc', 'p4012880.nc', 'p4012881.nc', 'p4012882.nc', 'p4012883.nc', 'p4012884.nc', 'p4012885.nc', 'p4012886.nc', 'p4012887.nc', 'p4012888.nc', 'p4012889.nc', 'p4012890.nc', 'p4012891.nc', 'p4012892.nc', 'p4012893.nc', 'p4012894.nc', 'p4012895.nc', 'p4012896.nc', 'p4012897.nc', 'p4012898.nc', 'p4012899.nc', 'p4012900.nc', 'p4012901.nc', 'p4012902.nc', 'p4012903.nc', 'p4012904.nc', 'p4012905.nc', 'p4012906.nc', 'p4012907.nc', 'p4012908.nc', 'p4012909.nc', 'p4012910.nc', 'p4012911.nc', 'p4012912.nc', 'p4012913.nc', 'p4012914.nc', 'p4012915.nc', 'p4012916.nc', 'p4012917.nc', 'p4012918.nc', 'p4012919.nc', 'p4012920.nc', 'p4012921.nc', 'p4012922.nc', 'p4012923.nc', 'p4012924.nc', 'p4012925.nc', 'p4012926.nc', 'p4012927.nc', 'p4012928.nc', 'p4012929.nc', 'p4012930.nc', 'p4012931.nc', 'p4012932.nc', 'p4012933.nc', 'p4012934.nc', 'p4012935.nc', 'p4012936.nc', 'p4012937.nc', 'p4012938.nc', 'p4012939.nc', 'p4012940.nc', 'p4012941.nc', 'p4012942.nc', 'p4012943.nc', 'p4012944.nc', 'p4012945.nc', 'p4012946.nc', 'p4012947.nc', 'p4012948.nc', 'p4012949.nc', 'p4012950.nc', 'p4012951.nc', 'p4012952.nc', 'p4012953.nc', 'p4012954.nc', 'p4012955.nc', 'p4012956.nc', 'p4012957.nc', 'p4012958.nc', 'p4012959.nc', 'p4012960.nc', 'p4012961.nc', 'p4012962.nc', 'p4012963.nc', 'p4012964.nc', 'p4012965.nc', 'p4012966.nc', 'p4012967.nc', 'p4012968.nc', 'p4012969.nc', 'p4012970.nc', 'p4012971.nc', 'p4012972.nc', 'p4012973.nc', 'p4012974.nc', 'p4012975.nc', 'p4012976.nc', 'p4012977.nc', 'p4012978.nc', 'p4012979.nc', 'p4012980.nc', 'p4012981.nc', 'p4012982.nc', 'p4012983.nc', 'p4012984.nc', 'p4012985.nc', 'p4012986.nc', 'p4012987.nc', 'p4012988.nc', 'p4012989.nc', 'p4012990.nc', 'p4012991.nc', 'p4012992.nc', 'p4012993.nc', 'p4012994.nc', 'p4012995.nc', 'p4012996.nc', 'p4012997.nc', 'p4012998.nc', 'p4012999.nc', 'p4013000.nc', 'p4013001.nc', 'p4013002.nc', 'p4013003.nc', 'p4013004.nc', 'p4013005.nc', 'p4013006.nc', 'p4013007.nc', 'p4013008.nc', 'p4013009.nc', 'p4013010.nc', 'p4013011.nc', 'p4013012.nc', 'p4013013.nc', 'p4013014.nc', 'p4013015.nc', 'p4013016.nc', 'p4013017.nc', 'p4013018.nc', 'p4013019.nc', 'p4013020.nc', 'p4013021.nc', 'p4013022.nc', 'p4013023.nc', 'p4013024.nc', 'p4013025.nc', 'p4013026.nc', 'p4013027.nc', 'p4013028.nc', 'p4013029.nc', 'p4013030.nc', 'p4013031.nc', 'p4013032.nc', 'p4013033.nc', 'p4013034.nc', 'p4013035.nc', 'p4013036.nc', 'p4013037.nc', 'p4013038.nc', 'p4013039.nc', 'p4013040.nc', 'p4013041.nc', 'p4013042.nc', 'p4013043.nc', 'p4013044.nc', 'p4013045.nc', 'p4013046.nc', 'p4013047.nc', 'p4013048.nc', 'p4013049.nc', 'p4013050.nc', 'p4013051.nc', 'p4013052.nc', 'p4013053.nc', 'p4013054.nc', 'p4013055.nc', 'p4013056.nc', 'p4013057.nc', 'p4013058.nc', 'p4013059.nc', 'p4013060.nc', 'p4013061.nc', 'p4013062.nc', 'p4013063.nc', 'p4013064.nc', 'p4013065.nc', 'p4013066.nc', 'p4013067.nc', 'p4013068.nc', 'p4013069.nc', 'p4013070.nc', 'p4013071.nc', 'p4013072.nc', 'p4013073.nc', 'p4013074.nc', 'p4013075.nc', 'p4013076.nc', 'p4013077.nc', 'p4013078.nc', 'p4013079.nc', 'p4013080.nc', 'p4013081.nc', 'p4013082.nc', 'p4013083.nc', 'p4013084.nc', 'p4013085.nc', 'p4013086.nc', 'p4013087.nc', 'p4013088.nc', 'p4013089.nc', 'p4013090.nc', 'p4013091.nc', 'p4013092.nc', 'p4013093.nc', 'p4013094.nc', 'p4013095.nc', 'p4013096.nc', 'p4013097.nc', 'p4013098.nc', 'p4013099.nc', 'p4013100.nc', 'p4013101.nc', 'p4013102.nc', 'p4013103.nc', 'p4013104.nc', 'p4013105.nc', 'p4013106.nc', 'p4013107.nc', 'p4013108.nc', 'p4013109.nc', 'p4013110.nc', 'p4013111.nc', 'p4013112.nc', 'p4013113.nc', 'p4013114.nc', 'p4013115.nc', 'p4013116.nc', 'p4013117.nc', 'p4013118.nc', 'p4013119.nc', 'p4013120.nc', 'p4013121.nc', 'p4013122.nc', 'p4013123.nc', 'p4013124.nc', 'p4013125.nc', 'p4013126.nc', 'p4013127.nc', 'p4013128.nc', 'p4013129.nc', 'p4013130.nc', 'p4013131.nc', 'p4013132.nc', 'p4013133.nc', 'p4013134.nc', 'p4013135.nc', 'p4013136.nc', 'p4013137.nc', 'p4013138.nc', 'p4013139.nc', 'p4013140.nc', 'p4013141.nc', 'p4013142.nc', 'p4013143.nc', 'p4013144.nc', 'p4013145.nc', 'p4013146.nc', 'p4013147.nc', 'p4013148.nc', 'p4013149.nc', 'p4013150.nc', 'p4013151.nc', 'p4013152.nc', 'p4013153.nc', 'p4013154.nc', 'p4013155.nc', 'p4013156.nc', 'p4013157.nc', 'p4013158.nc', 'p4013159.nc', 'p4013160.nc', 'p4013161.nc', 'p4013162.nc', 'p4013163.nc', 'p4013164.nc', 'p4013165.nc', 'p4013166.nc', 'p4013167.nc', 'p4013168.nc', 'p4013169.nc', 'p4013170.nc', 'p4013171.nc', 'p4013172.nc', 'p4013173.nc', 'p4013174.nc', 'p4013175.nc', 'p4013176.nc', 'p4013177.nc', 'p4013178.nc', 'p4013179.nc', 'p4013180.nc', 'p4013181.nc', 'p4013182.nc', 'p4013183.nc', 'p4013184.nc', 'p4013185.nc', 'p4013186.nc', 'p4013187.nc', 'p4013188.nc', 'p4013189.nc', 'p4013190.nc', 'p4013191.nc', 'p4013192.nc', 'p4013193.nc', 'p4013194.nc', 'p4013195.nc', 'p4013196.nc', 'p4013197.nc', 'p4013198.nc', 'p4013199.nc', 'p4013200.nc', 'p4013201.nc', 'p4013202.nc', 'p4013203.nc', 'p4013204.nc', 'p4013205.nc', 'p4013206.nc', 'p4013207.nc', 'p4013208.nc', 'p4013209.nc', 'p4013210.nc', 'p4013211.nc', 'p4013212.nc', 'p4013213.nc', 'p4013214.nc', 'p4013215.nc', 'p4013216.nc', 'p4013217.nc', 'p4013218.nc', 'p4013219.nc', 'p4013220.nc', 'p4013221.nc', 'p4013222.nc', 'p4013223.nc', 'p4013224.nc', 'p4013225.nc', 'p4013226.nc', 'p4013227.nc', 'p4013228.nc', 'p4013229.nc', 'p4013230.nc', 'p4013231.nc', 'p4013232.nc', 'p4013233.nc', 'p4013234.nc', 'p4013235.nc', 'p4013236.nc', 'p4013237.nc', 'p4013238.nc', 'p4013239.nc', 'p4013240.nc', 'p4013241.nc', 'p4013242.nc', 'p4013243.nc', 'p4013244.nc', 'p4013245.nc', 'p4013246.nc', 'p4013247.nc', 'p4013248.nc', 'p4013249.nc', 'p4013250.nc', 'p4013251.nc', 'p4013252.nc', 'p4013253.nc', 'p4013254.nc', 'p4013255.nc', 'p4013256.nc', 'p4013257.nc', 'p4013258.nc', 'p4013259.nc', 'p4013260.nc', 'p4013261.nc', 'p4013263.nc', 'p4013264.nc', 'p4013265.nc', 'p4013266.nc', 'p4013267.nc', 'p4013268.nc', 'p4013269.nc', 'p4013270.nc', 'p4013271.nc', 'p4013272.nc', 'p4013273.nc', 'p4013274.nc', 'p4013275.nc', 'p4013276.nc', 'p4013277.nc', 'p4013278.nc', 'p4013279.nc', 'p4013280.nc', 'p4013281.nc', 'p4013282.nc', 'p4013283.nc', 'p4013284.nc', 'p4013285.nc', 'p4013286.nc', 'p4013287.nc', 'p4013288.nc', 'p4013289.nc', 'p4013290.nc', 'p4013291.nc', 'p4013292.nc', 'p4013293.nc', 'p4013294.nc', 'p4013295.nc', 'p4013296.nc', 'p4013297.nc', 'p4013298.nc', 'p4013299.nc', 'p4013300.nc', 'p4013301.nc', 'p4013302.nc', 'p4013303.nc', 'p4013304.nc', 'p4013305.nc', 'p4013306.nc', 'p4013307.nc', 'p4013308.nc', 'p4013309.nc', 'p4013310.nc', 'p4013311.nc', 'p4013312.nc', 'p4013313.nc', 'p4013314.nc', 'p4013315.nc', 'p4013316.nc', 'p4013317.nc', 'p4013318.nc', 'p4013319.nc', 'p4013320.nc', 'p4013321.nc', 'p4013322.nc', 'p4013323.nc', 'p4013324.nc', 'p4013325.nc', 'p4013326.nc', 'p4013327.nc', 'p4013328.nc', 'p4013329.nc', 'p4013330.nc', 'p4013331.nc', 'p4013332.nc', 'p4013333.nc', 'p4013334.nc', 'p4013335.nc', 'p4013336.nc', 'p4013337.nc', 'p4013338.nc', 'p4013339.nc', 'p4013340.nc', 'p4013341.nc', 'p4013342.nc', 'p4013343.nc', 'p4013344.nc', 'p4013345.nc', 'p4013346.nc', 'p4013347.nc', 'p4013348.nc', 'p4013349.nc', 'p4013350.nc', 'p4013351.nc', 'p4013352.nc', 'p4013353.nc', 'p4013354.nc', 'p4013355.nc', 'p4013356.nc', 'p4013357.nc', 'p4013358.nc', 'p4013359.nc', 'p4013360.nc', 'p4013361.nc', 'p4013362.nc', 'p4013363.nc', 'p4013364.nc', 'p4013365.nc', 'p4013366.nc', 'p4013367.nc', 'p4013368.nc', 'p4013369.nc', 'p4013370.nc', 'p4013371.nc', 'p4013372.nc', 'p4013373.nc', 'p4013374.nc', 'p4013375.nc', 'p4013376.nc', 'p4013377.nc', 'p4013378.nc', 'p4013379.nc', 'p4013380.nc', 'p4013381.nc', 'p4013382.nc', 'p4013383.nc', 'p4013384.nc', 'p4013385.nc', 'p4013386.nc', 'p4013387.nc', 'p4013388.nc', 'p4013389.nc', 'p4013390.nc', 'p4013391.nc', 'p4013392.nc', 'p4013393.nc', 'p4013394.nc', 'p4013395.nc', 'p4013396.nc', 'p4013397.nc', 'p4013398.nc', 'p4013399.nc', 'p4013400.nc', 'p4013401.nc', 'p4013402.nc', 'p4013403.nc', 'p4013404.nc', 'p4013405.nc', 'p4013406.nc', 'p4013407.nc', 'p4013408.nc', 'p4013409.nc', 'p4013410.nc', 'p4013411.nc', 'p4013412.nc', 'p4013413.nc', 'p4013414.nc', 'p4013415.nc', 'p4013416.nc', 'p4013417.nc', 'p4013418.nc', 'p4013419.nc', 'p4013420.nc', 'p4013421.nc', 'p4013422.nc', 'p4013423.nc', 'p4013424.nc', 'p4013425.nc', 'p4013426.nc', 'p4013427.nc', 'p4013428.nc', 'p4013429.nc', 'p4013430.nc', 'p4013431.nc', 'p4013432.nc', 'p4013433.nc', 'p4013434.nc', 'p4013435.nc', 'p4013436.nc', 'p4013437.nc', 'p4013438.nc', 'p4013439.nc', 'p4013440.nc', 'p4013441.nc', 'p4013442.nc', 'p4013443.nc', 'p4013444.nc', 'p4013445.nc', 'p4013446.nc', 'p4013447.nc', 'p4013448.nc', 'p4013449.nc', 'p4013450.nc', 'p4013451.nc', 'p4013452.nc', 'p4013453.nc', 'p4013454.nc', 'p4013455.nc', 'p4013456.nc', 'p4013457.nc', 'p4013458.nc', 'p4013459.nc', 'p4013460.nc', 'p4013461.nc', 'p4013462.nc', 'p4013464.nc', 'p4013465.nc', 'p4013466.nc', 'p4013467.nc', 'p4013468.nc', 'p4013469.nc', 'p4013470.nc', 'p4013471.nc', 'p4013472.nc', 'p4013473.nc', 'p4013474.nc', 'p4013475.nc', 'p4013476.nc', 'p4013477.nc', 'p4013478.nc', 'p4013479.nc', 'p4013480.nc', 'p4013481.nc', 'p4013482.nc', 'p4013483.nc', 'p4013484.nc', 'p4013485.nc', 'p4013486.nc', 'p4013487.nc', 'p4013488.nc', 'p4013489.nc', 'p4013490.nc', 'p4013491.nc', 'p4013492.nc', 'p4013493.nc', 'p4013494.nc', 'p4013495.nc', 'p4013496.nc', 'p4013497.nc', 'p4013498.nc', 'p4013499.nc', 'p4013500.nc', 'p4013501.nc', 'p4013502.nc', 'p4013503.nc', 'p4013504.nc', 'p4013505.nc', 'p4013506.nc', 'p4013507.nc', 'p4013508.nc', 'p4013509.nc', 'p4013510.nc', 'p4013511.nc', 'p4013512.nc', 'p4013513.nc', 'p4013514.nc', 'p4013515.nc', 'p4013516.nc', 'p4013517.nc', 'p4013518.nc', 'p4013519.nc', 'p4013520.nc', 'p4013521.nc', 'p4013522.nc', 'p4013523.nc', 'p4013524.nc', 'p4013525.nc', 'p4013526.nc', 'p4013527.nc', 'p4013528.nc', 'p4013529.nc', 'p4013530.nc', 'p4013531.nc', 'p4013532.nc', 'p4013533.nc', 'p4013534.nc', 'p4013535.nc', 'p4013536.nc', 'p4013537.nc', 'p4013538.nc', 'p4013539.nc', 'p4013540.nc', 'p4013541.nc', 'p4013542.nc', 'p4013543.nc', 'p4013544.nc', 'p4013545.nc', 'p4013546.nc', 'p4013547.nc', 'p4013548.nc', 'p4013549.nc', 'p4013550.nc', 'p4013551.nc', 'p4013552.nc', 'p4013553.nc', 'p4013554.nc', 'p4013555.nc', 'p4013556.nc', 'p4013557.nc', 'p4013558.nc', 'p4013559.nc', 'p4013560.nc', 'p4013561.nc', 'p4013562.nc', 'p4013563.nc', 'p4013564.nc', 'p4013565.nc', 'p4013566.nc', 'p4013567.nc', 'p4013568.nc', 'p4013569.nc', 'p4013570.nc', 'p4013571.nc', 'p4013572.nc', 'p4013573.nc', 'p4013574.nc', 'p4013575.nc', 'p4013576.nc', 'p4013577.nc', 'p4013578.nc', 'p4013579.nc', 'p4013580.nc', 'p4013581.nc', 'p4013582.nc', 'p4013583.nc', 'p4013584.nc', 'p4013585.nc', 'p4013586.nc', 'p4013587.nc', 'p4013588.nc', 'p4013589.nc', 'p4013590.nc', 'p4013591.nc', 'p4013592.nc', 'p4013593.nc', 'p4013594.nc', 'p4013595.nc', 'p4013596.nc', 'p4013597.nc', 'p4013598.nc', 'p4013599.nc', 'p4013600.nc', 'p4013601.nc', 'p4013602.nc', 'p4013603.nc', 'p4013604.nc', 'p4013605.nc', 'p4013606.nc', 'p4013607.nc', 'p4013608.nc', 'p4013609.nc', 'p4013610.nc', 'p4013611.nc', 'p4013612.nc']
- time_coverage_start :
- 2017-08-13 21:07:40
- time_coverage_end :
- 2017-09-26 16:05:42
- geospatial_vertical_min :
- -0.5030842835078432
- geospatial_vertical_max :
- 67.57316616647842
- geospatial_lat_min :
- 57.89913235353872
- geospatial_lat_max :
- 62.14208166934556
- geospatial_lon_min :
- -174.5055983333333
- geospatial_lon_max :
- -168.8639759711956
- processing :
- [2021-07-26 13:55:28] imported data with GliderTools.load.seaglider_basestation_netCDFs;
dat = ds_dict['sg_data_point'].rename({
'ctd_pressure': 'pressure',
'ctd_depth': 'depth',
'ctd_time_dt64': 'time',
'eng_wlbb2fl_sig695nm': 'flr_raw'
})
# name coordinates for quicker plotting
x = dat.dives
y = dat.depth
gt.plot(x, y, dat.temperature_raw, shading='nearest', cmap=cmo.thermal, robust=True)
title('Original Data')
show()
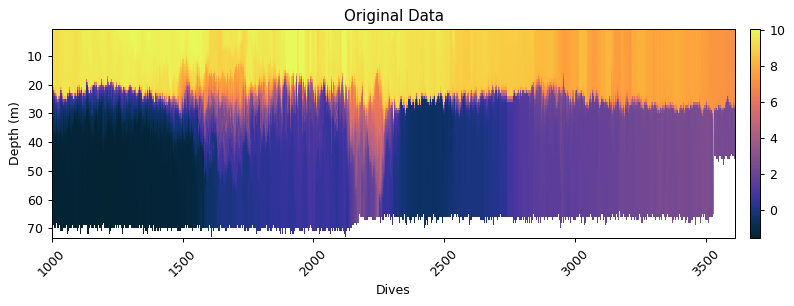
gt.plot(x, y, dat.temperature, shading='nearest', cmap=cmo.thermal, robust=True)
title('Original Data - UW QC')
show()
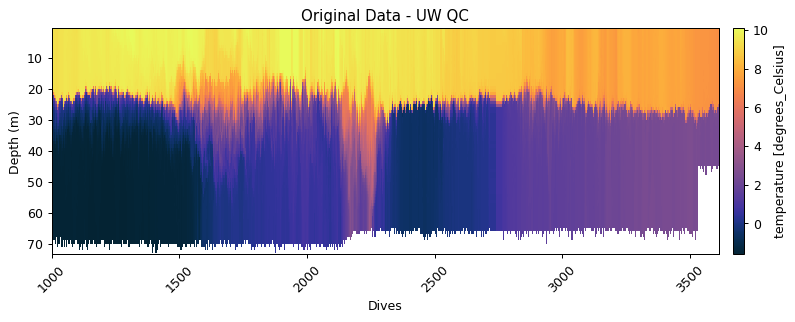
#lots of options available, but lets filter (without much smoothing if any at all, lets do it in the verticle for sure and contemplate horizonatl filtering too)
temp_qc = gt.calc_physics(dat.temperature, x, y,
iqr=1.5, depth_threshold=0, #<-- depth
spike_window=5, spike_method='median',
savitzky_golay_window=0, savitzky_golay_order=2) #<--challenged by sharp interface, made it clearly wrong with window=11, order=2
# PLOTTING
fig, ax = plt.subplots(3, 1, figsize=[9, 8.5], sharex=True, dpi=90)
gt.plot(x, y, dat.temperature, cmap=cmo.thermal, ax=ax[0])
gt.plot(x, y, temp_qc, cmap=cmo.thermal, ax=ax[1])
gt.plot(x, y, temp_qc - dat.temperature, cmap=cm.RdBu_r, vmin=-0.05, vmax=0.05, ax=ax[2])
[a.set_xlabel('') for a in ax]
ax[0].cb.set_label('Original Data')
ax[1].cb.set_label('Cleaned Data')
ax[2].cb.set_label('Difference from Original')
plt.show()
==================================================
Physics Variable:
Removing outliers with IQR * 1.5: 0 obs
Removing spikes with rolling median (spike window=5)
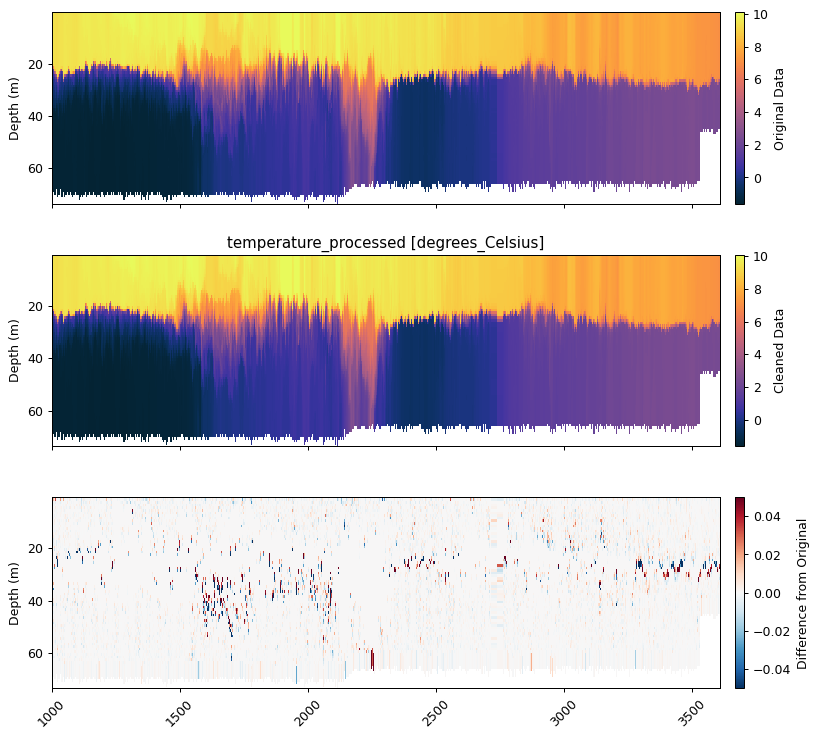
#lots of options available, but lets filter (without much smoothing if any at all, lets do it in the verticle for sure and contemplate horizonatl filtering too)
# how does it do for despiking at interface?
salt_qc = gt.calc_physics(dat.salinity, x, y,
mask_frac=0.2, iqr=2.5,
spike_window=5, spike_method='median',
savitzky_golay_window=0, savitzky_golay_order=2)
# PLOTTING
fig, ax = plt.subplots(3, 1, figsize=[9, 8.5], sharex=True, dpi=90)
gt.plot(x, y, dat.salinity, cmap=cmo.haline, ax=ax[0])
gt.plot(x, y, salt_qc, cmap=cmo.haline, ax=ax[1])
gt.plot(x, y, salt_qc - dat.salinity, cmap=cm.RdBu_r, vmin=-0.02, vmax=0.02, ax=ax[2])
[a.set_xlabel('') for a in ax]
ax[0].cb.set_label('Original Data')
ax[1].cb.set_label('Cleaned Data')
ax[2].cb.set_label('Difference from Original')
plt.show()
==================================================
Physics Variable:
Removing outliers with IQR * 2.5: 313 obs
Removing spikes with rolling median (spike window=5)
Removing horizontal outliers (fraction=0.2, multiplier=2.5)
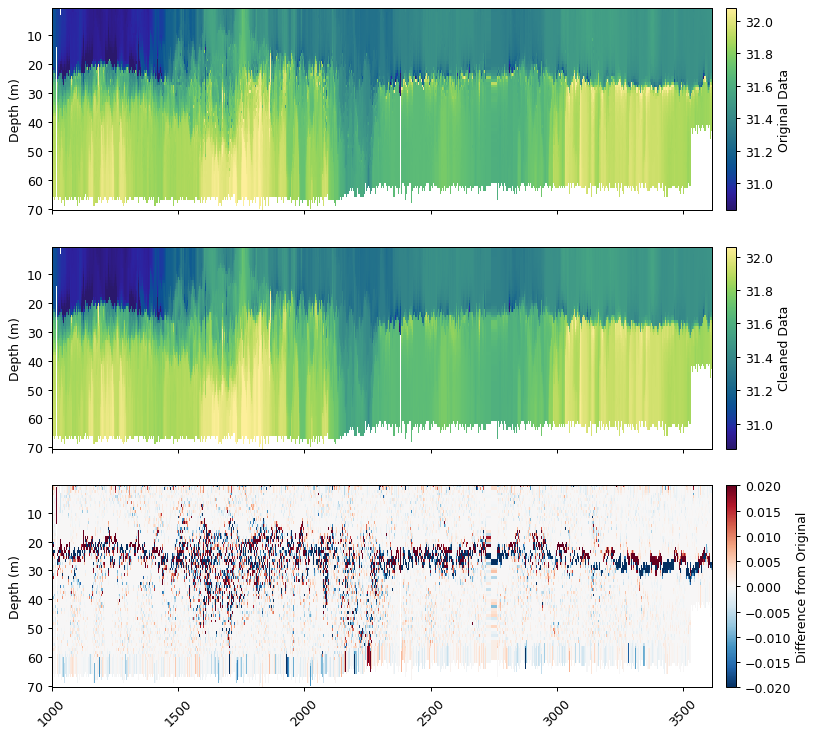
Insert data into original dataset¶
dat['temp_gt_qc'] = temp_qc
dat['salt_gt_qc'] = salt_qc
Derived Variables¶
dens0 = gt.physics.potential_density(salt_qc, temp_qc, dat.pressure, dat.latitude, dat.longitude)
mld = gt.physics.mixed_layer_depth(x, y, dens0)
mld_smoothed = mld.rolling(10, min_periods=3).mean()
mld_mask = gt.physics.mixed_layer_depth(x, y, dens0, return_as_mask=True)
mld_grid = gt.grid_data(x, y, mld_mask, verbose=False)
fig, ax = plt.subplots(1, 2, figsize=[9, 3], dpi=100, sharey=True)
mld_smoothed.plot(ax=ax[0])
gt.plot(mld_grid, ax=ax[1])
[a.set_ylim(100, 0) for a in ax]
ax[0].set_ylabel('Depth (m)')
[a.set_xlabel('Dives') for a in ax]
xticks(rotation=0)
fig.tight_layout()
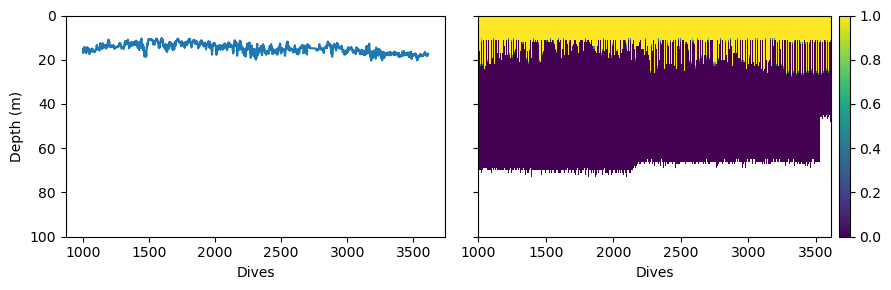
Ancillary Instrumentation: Backscatter/Fluorometry , Oxygen , PAR¶
apply calibration curves from factor
despike in vertical
quenching and other fluorometric adjustments
oxygen equilibration in ML boundary
Gridding and interpolation¶
Vertical gridding¶
It is often more convenient and computationally efficient to work with data that has been gridded to a standard vertical grid (i.e. depths have been binned). GliderTools offers very easy to use and efficient tools to grid data once all the processing has been completed.
The first task is to select the bin size of the data that will be gridded. GliderTools automatically selects bin sizes according to the sampling frequency of the dataset for every 50m. This is shown in the figure below, where the 2D histogram shows the sampling frequency (by depth) and the line shows the automatically selected bin size rounded up to the nearest 0.5m.
ax = gt.plot.bin_size(dat.depth, cmap=mpl.cm.Blues)
ax.set_xlim(0, 6)
line = ax.get_children()[1]
line.set_linewidth(6)
line.set_color('orange')
legend = ax.get_children()[-2]
legend.set_visible(False)
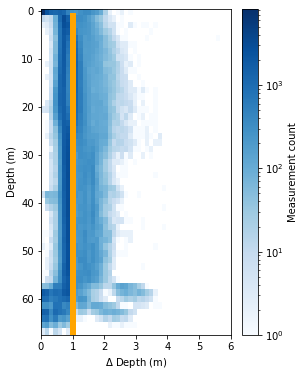
1m seems the optimal gridding for the 2017 SG missions
